
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,905,884 – 8,905,991 |
| Length | 107 |
| Max. P | 0.996773 |

| Location | 8,905,884 – 8,905,991 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.51008 |
| G+C content | 0.38068 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -13.35 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
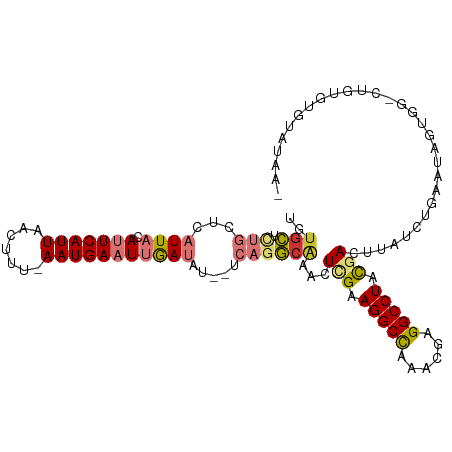
>dm3.chr2R 8905884 107 + 21146708 UGUGCUCUGCUCAUUACAUUCAUUAACUUU-AAUGAAUUGAUAUAUUCAGGCAAACUCGAAGGCCAUCAAAGGCCUACGACUUAUCUGAAUAUUGG-CUGUGUGUAUAA- (((((.(.(((.((((.((((((((....)-)))))))))))((((((((..((..(((.(((((......))))).))).))..)))))))).))-)...).))))).- ( -31.90, z-score = -4.11, R) >dp4.chr3 5675892 102 + 19779522 UGUGUGCUGCUCAUUACAGUCAUUGACUCG-AAUGAAU-AACGC-----AACGAAUUCGAAGGCUAAAC-AGGCCUAUAAAUUGCCUGAAUAGUUGCCUGUGAAUCGAAA (((((.....(((((..((((...))))..-)))))..-.))))-----).(((.((((.((((....(-((((.........))))).......)))).)))))))... ( -27.10, z-score = -2.44, R) >droPer1.super_2 5868974 87 + 9036312 --UGUG-UGCUCAUUACAGUCAUUGACUCG-AAUGAAU-AACGC-----AACGAAUUCGAAGGCUAAAC-AGGCCUAUAAAUUGC------------CUGUGAAUCGAAA --((((-(..(((((..((((...))))..-)))))..-.))))-----).....(((((.......((-((((.........))------------))))...))))). ( -22.10, z-score = -2.32, R) >droAna3.scaffold_13266 6378187 106 + 19884421 UGUGUUUUGCUCAUUACAUUCAUUAACUUUUAAUGAAUUGAUAU--UCAGAGAA-CUUGAAGGCUGAAUGAGGCCUACGACUUGCUUGAAAAGUAG-CUGAUUUUCAAAA .((.(((((...((((.((((((((.....))))))))))))..--.))))).)-)(((.(((((......))))).))).....(((((((....-....))))))).. ( -20.60, z-score = -0.54, R) >droEre2.scaffold_4845 5675676 105 - 22589142 UUUGCUCUGCUCAUUACAUUCAUUAGGUUU-AAUGAAUUGAUAU--UCAGGCAA-CUCGAAGGCCGUCAGAGGCCUACGACUUAUCUGAAAAGUGG-AUGUGCGUAUAAU ...((((..((.((((.((((((((....)-))))))))))).(--((((..((-.(((.(((((......))))).))).))..))))).))..)-)...))....... ( -31.10, z-score = -2.67, R) >droYak2.chr2R 13082003 104 - 21139217 UGUGCUCUGCUCAUUACAUUCAUUAAGUUU-AAUGAAUUGAUAU--UCAGGCAAACUCGAAGGCCAUCCGAGGCCUACGACUUAUAAAAAAAGUG---UAUGUGUAUAAU (((((.(((...((((.((((((((....)-)))))))))))..--.)))(((.((.((.(((((......))))).))((((.......)))))---).)))))))).. ( -24.10, z-score = -1.93, R) >droSec1.super_1 6422085 105 + 14215200 UGCGCUCUGCUCAUUACAUUCAUUAACUUU-AAUGAAUUGAUAU--UCAGGCAAACUCGAAGGCCAACUGAGGCCUACGACUUAUCUGAAUAUUGG-CUGUGUGCUUAA- .((((.(.(((......((((((((....)-))))))).(((((--((((..((..(((.(((((......))))).))).))..)))))))))))-).).))))....- ( -35.90, z-score = -5.51, R) >droSim1.chr2R 7357188 105 + 19596830 UGUGCUCUGCUCAUUACAUUCAUUAACUUU-AAUGAAUUGAUAU--UCAGGCAAACUCGAAGGCCAAAUGAGGCCUACGACUUAUCUGAAUAUUGG-CUGUGUGUAUAG- (((((.(.(((......((((((((....)-))))))).(((((--((((..((..(((.(((((......))))).))).))..)))))))))))-)...).))))).- ( -32.70, z-score = -4.19, R) >consensus UGUGCUCUGCUCAUUACAUUCAUUAACUUU_AAUGAAUUGAUAU__UCAGGCAAACUCGAAGGCCAAACGAGGCCUACGACUUAUCUGAAUAGUGG_CUGUGUGUAUAA_ ..((((...........(((((((.......)))))))...........))))...(((.(((((......))))).))).............................. (-13.35 = -13.22 + -0.12)
| Location | 8,905,884 – 8,905,991 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.51008 |
| G+C content | 0.38068 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
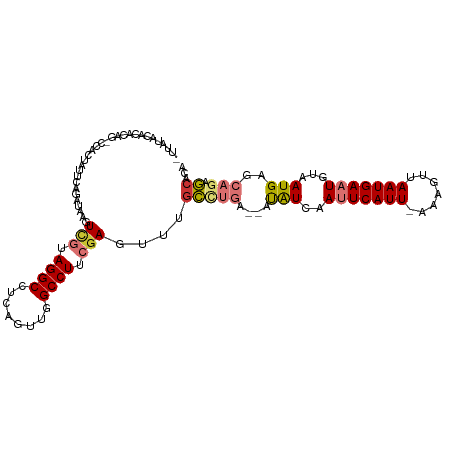
>dm3.chr2R 8905884 107 - 21146708 -UUAUACACACAG-CCAAUAUUCAGAUAAGUCGUAGGCCUUUGAUGGCCUUCGAGUUUGCCUGAAUAUAUCAAUUCAUU-AAAGUUAAUGAAUGUAAUGAGCAGAGCACA -...........(-(((((((((((.(((((((.(((((......))))).))).)))).))))))))....(((((((-(....))))))))....)).))........ ( -30.80, z-score = -3.90, R) >dp4.chr3 5675892 102 - 19779522 UUUCGAUUCACAGGCAACUAUUCAGGCAAUUUAUAGGCCU-GUUUAGCCUUCGAAUUCGUU-----GCGUU-AUUCAUU-CGAGUCAAUGACUGUAAUGAGCAGCACACA .(((((.....((((...((..(((((.........))))-)..)))))))))))...(((-----((...-..(((((-..((((...))))..))))))))))..... ( -26.90, z-score = -2.40, R) >droPer1.super_2 5868974 87 - 9036312 UUUCGAUUCACAG------------GCAAUUUAUAGGCCU-GUUUAGCCUUCGAAUUCGUU-----GCGUU-AUUCAUU-CGAGUCAAUGACUGUAAUGAGCA-CACA-- .(((((...((((------------((.........))))-)).......)))))((((((-----(((((-(((.((.-...)).)))))).))))))))..-....-- ( -21.60, z-score = -1.97, R) >droAna3.scaffold_13266 6378187 106 - 19884421 UUUUGAAAAUCAG-CUACUUUUCAAGCAAGUCGUAGGCCUCAUUCAGCCUUCAAG-UUCUCUGA--AUAUCAAUUCAUUAAAAGUUAAUGAAUGUAAUGAGCAAAACACA (((((....(((.-.(((..((((((.((.(.(.((((........)))).).).-)))).)))--).....((((((((.....))))))))))).))).))))).... ( -17.30, z-score = -0.67, R) >droEre2.scaffold_4845 5675676 105 + 22589142 AUUAUACGCACAU-CCACUUUUCAGAUAAGUCGUAGGCCUCUGACGGCCUUCGAG-UUGCCUGA--AUAUCAAUUCAUU-AAACCUAAUGAAUGUAAUGAGCAGAGCAAA .......((.(((-..((..(((((.(((.(((.(((((......))))).))).-))).))))--).....(((((((-(....)))))))))).))).))........ ( -29.80, z-score = -3.59, R) >droYak2.chr2R 13082003 104 + 21139217 AUUAUACACAUA---CACUUUUUUUAUAAGUCGUAGGCCUCGGAUGGCCUUCGAGUUUGCCUGA--AUAUCAAUUCAUU-AAACUUAAUGAAUGUAAUGAGCAGAGCACA ............---............((.(((.(((((......))))).))).))((((((.--.(((..(((((((-(....))))))))...)))..))).))).. ( -26.20, z-score = -2.84, R) >droSec1.super_1 6422085 105 - 14215200 -UUAAGCACACAG-CCAAUAUUCAGAUAAGUCGUAGGCCUCAGUUGGCCUUCGAGUUUGCCUGA--AUAUCAAUUCAUU-AAAGUUAAUGAAUGUAAUGAGCAGAGCGCA -....((.(.(.(-(((((((((((.(((((((.(((((......))))).))).)))).))))--))))..(((((((-(....))))))))....)).)).).).)). ( -32.50, z-score = -4.04, R) >droSim1.chr2R 7357188 105 - 19596830 -CUAUACACACAG-CCAAUAUUCAGAUAAGUCGUAGGCCUCAUUUGGCCUUCGAGUUUGCCUGA--AUAUCAAUUCAUU-AAAGUUAAUGAAUGUAAUGAGCAGAGCACA -...........(-(((((((((((.(((((((.(((((......))))).))).)))).))))--))))..(((((((-(....))))))))....)).))........ ( -30.20, z-score = -3.84, R) >consensus _UUAUACACACAG_CCACUAUUCAGAUAAGUCGUAGGCCUCAGUUGGCCUUCGAGUUUGCCUGA__AUAUCAAUUCAUU_AAAGUUAAUGAAUGUAAUGAGCAGAGCACA ..............................(((.((((........)))).)))....(((((.........(((((((.......)))))))........))).))... (-12.15 = -12.94 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:07 2011