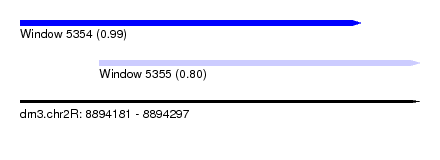
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,894,181 – 8,894,297 |
| Length | 116 |
| Max. P | 0.985942 |

| Location | 8,894,181 – 8,894,280 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Shannon entropy | 0.40540 |
| G+C content | 0.46014 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.19 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
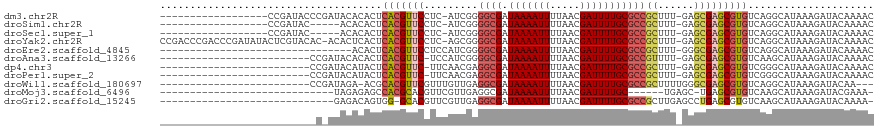
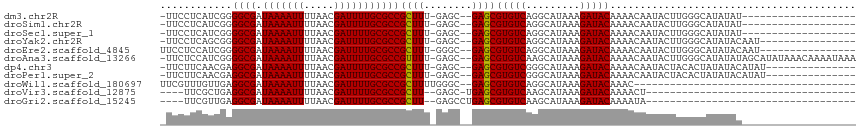
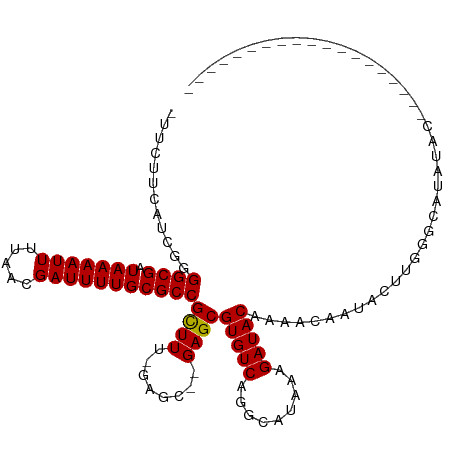
>dm3.chr2R 8894181 99 + 21146708 ------------------CCGAUACCCGAUACACACUCACGUUCCUC-AUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCAGGCAUAAAGAUACAAAAC ------------------......((.((........((((((((((-(..(.((((.(((((((.....))))))))))).)..)-))).))))))))).))................ ( -28.40, z-score = -2.60, R) >droSim1.chr2R 7344855 94 + 19596830 ------------------CCGAUAC-----ACACACUCACGUUCCUC-AUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCAGGCAUAAAGAUACAAAAC ------------------.......-----.......((((((((((-(..(.((((.(((((((.....))))))))))).)..)-))).)))))))..................... ( -28.10, z-score = -3.11, R) >droSec1.super_1 6410402 94 + 14215200 ------------------CCGAUAC-----ACACACUCACGUUCCUC-AUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCAGGCAUAAAGAUACAAAAC ------------------.......-----.......((((((((((-(..(.((((.(((((((.....))))))))))).)..)-))).)))))))..................... ( -28.10, z-score = -3.11, R) >droYak2.chr2R 13069902 116 - 21139217 CCGACCCGACCCGAUAUACUCGUACAC-ACACUCACUCACGUUCCUC-AGCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCAGGCAUAAAGAUACAAAAC ..(.((.(((.(((.....))).....-..........(((((((((-((.(.((((.(((((((.....))))))))))).).))-))).))))))))).)))............... ( -32.00, z-score = -2.26, R) >droEre2.scaffold_4845 5663544 86 - 22589142 --------------------------------ACACUCACGUUCCUCCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GGGCGAGCGUGUCAGGCAUAAAGAUACAAAAC --------------------------------.....(((((((.((((..(.((((.(((((((.....))))))))))).)..)-))).)))))))..................... ( -27.80, z-score = -2.96, R) >droAna3.scaffold_13266 6366173 92 + 19884421 -------------------------CCGAUACACACUCACGUUC-UCCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGUUUU-GAGCGAGCGUGUCAAGCAUAAAGAUACAAAAC -------------------------............(((((((-.(((..(.((((.(((((((.....))))))))))).)..)-).).)))))))..................... ( -22.20, z-score = -1.35, R) >dp4.chr3 5663414 92 + 19779522 -------------------------CCGAUACAUACUCACGUUC-UUCAACGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCGGGCAUAAAGAUACAAAAC -------------------------((((........(((((((-(((((.(.((((.(((((((.....))))))))))).).))-))).)))))))))))................. ( -28.10, z-score = -3.07, R) >droPer1.super_2 5857847 92 + 9036312 -------------------------CCGAUACAUACUCACGUUC-UUCAACGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGCGAGCGUGUCGGGCAUAAAGAUACAAAAC -------------------------((((........(((((((-(((((.(.((((.(((((((.....))))))))))).).))-))).)))))))))))................. ( -28.10, z-score = -3.07, R) >droWil1.scaffold_180697 3233311 90 - 4168966 -------------------------CCGAUAGA-ACGCACGUUCGUUUGUUGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUUGGGCGAGCGUGUCAGGCAUAAAGAUACAA--- -------------------------........-..((((((((((..(..(.((((.(((((((.....))))))))))).)...)..)))))))))).................--- ( -28.00, z-score = -1.79, R) >droMoj3.scaffold_6496 3653464 82 + 26866924 -----------------------------UAGAGAGCCACGCACGUUCGUUGAGGCGAUAAAAUUUUAACGAUUUUGC------UGAGC-UGAGCGUGUCAAGCAUAAAGAUACGAAA- -----------------------------.....(((((.(((...(((((((((........)))))))))...)))------)).))-)...((((((.........))))))...- ( -20.40, z-score = -1.04, R) >droGri2.scaffold_15245 8008060 88 - 18325388 -----------------------------GAGACAGUGG-GCACGUUCGUUGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUGAGCCUGAGCGUGUCAAGCAUAAAGAUACAAAA- -----------------------------......((.(-((((((((.....((((.(((((((.....)))))))))))((....))..)))))))))..))..............- ( -26.10, z-score = -1.74, R) >consensus __________________________C_AUACACACUCACGUUCCUCCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU_GAGCGAGCGUGUCAGGCAUAAAGAUACAAAAC .....................................(((((((.........((((.(((((((.....)))))))))))(((....))))))))))..................... (-17.65 = -18.19 + 0.54)
| Location | 8,894,204 – 8,894,297 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.39954 |
| G+C content | 0.40779 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
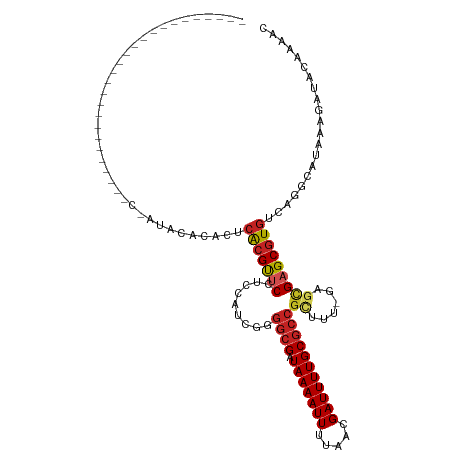
>dm3.chr2R 8894204 93 + 21146708 -UUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUAU------------------- -.((((((..(.((((.(((((((.....))))))))))).)..)-))).--))(((((((.........)))))....((.....)).))......------------------- ( -23.20, z-score = -1.24, R) >droSim1.chr2R 7344873 93 + 19596830 -UUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUAU------------------- -.((((((..(.((((.(((((((.....))))))))))).)..)-))).--))(((((((.........)))))....((.....)).))......------------------- ( -23.20, z-score = -1.24, R) >droSec1.super_1 6410420 93 + 14215200 -UUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUAU------------------- -.((((((..(.((((.(((((((.....))))))))))).)..)-))).--))(((((((.........)))))....((.....)).))......------------------- ( -23.20, z-score = -1.24, R) >droYak2.chr2R 13069942 96 - 21139217 -UUCCUCAGCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUACAAU---------------- -.(((((((.(.((((.(((((((.....))))))))))).).))-))).--))(((((((.........)))))....((.....)).)).........---------------- ( -23.70, z-score = -1.14, R) >droEre2.scaffold_4845 5663553 97 - 22589142 UUCCUCCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GGGC--GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUACAAU---------------- .((.((((..(.((((.(((((((.....))))))))))).)..)-))).--))(((((((.........)))))....((.....)).)).........---------------- ( -22.90, z-score = -0.68, R) >droAna3.scaffold_13266 6366189 112 + 19884421 -UUCUCCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGUUUU-GAGC--GAGCGUGUCAAGCAUAAAGAUACAAAACAAUACUUGGGCAUAUAUAGCAUAUAAACAAAAUAAA -....(((....((((.(((((((.....)))))))))))(((((-(..(--....(((.....)))...)...))))))......)))((.......))................ ( -18.90, z-score = 0.32, R) >dp4.chr3 5663430 97 + 19779522 -UUCUUCAACGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCGGGCAUAAAGAUACAAAACAAUACUACACUAUAUACAUAU--------------- -((((((((.(.((((.(((((((.....))))))))))).).))-))).--))).(((((.........)))))..........................--------------- ( -19.50, z-score = -1.18, R) >droPer1.super_2 5857863 97 + 9036312 -UUCUUCAACGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU-GAGC--GAGCGUGUCGGGCAUAAAGAUACAAAACAAUACUACACUAUAUACAUAU--------------- -((((((((.(.((((.(((((((.....))))))))))).).))-))).--))).(((((.........)))))..........................--------------- ( -19.50, z-score = -1.18, R) >droWil1.scaffold_180697 3233326 77 - 4168966 UUCGUUUGUUGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUUGGGC--GAGCGUGUCAGGCAUAAAGAUACAAAC------------------------------------- (((((..(..(.((((.(((((((.....))))))))))).)...)..))--))).(((((.........)))))....------------------------------------- ( -20.50, z-score = -1.14, R) >droVir3.scaffold_12875 644837 74 + 20611582 ----UUCGCUGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUU--GAGC-UGAGCGUGUCAAGCAUAAAGAUACAAAACU----------------------------------- ----...(((.(((((.(((((((.....)))))))))))((..--..))-).)))(((((.........)))))......----------------------------------- ( -19.10, z-score = -1.11, R) >droGri2.scaffold_15245 8008075 75 - 18325388 ----UUCGUUGAGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUU--GAGCCUGAGCGUGUCAAGCAUAAAGAUACAAAAUA----------------------------------- ----....(((.((((.(((((((.....)))))))))))((((--((((....))...)))))).........)))....----------------------------------- ( -18.80, z-score = -1.34, R) >consensus _UUCUUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUU_GAGC__GAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUAC___________________ ............((((.(((((((.....)))))))))))((((........))))(((((.........)))))......................................... (-17.48 = -17.40 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:04 2011