| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,808,733 – 8,808,849 |
| Length | 116 |
| Max. P | 0.998878 |

| Location | 8,808,733 – 8,808,849 |
|---|---|
| Length | 116 |
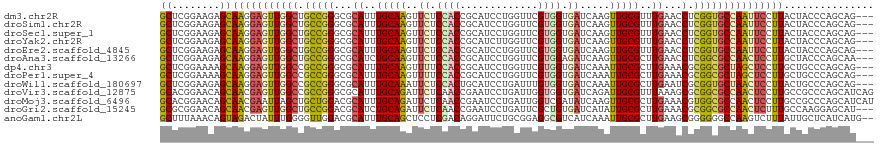
| Sequences | 13 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Shannon entropy | 0.38026 |
| G+C content | 0.55848 |
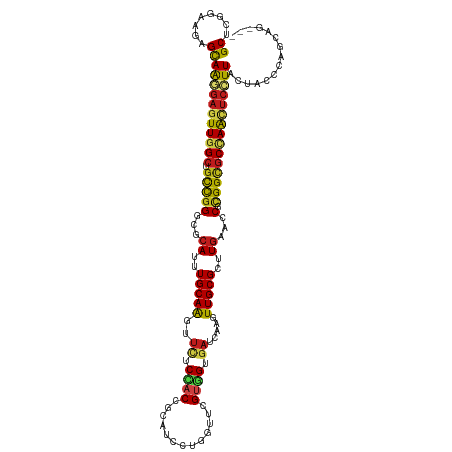
| Mean single sequence MFE | -44.89 |
| Consensus MFE | -19.84 |
| Energy contribution | -19.89 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
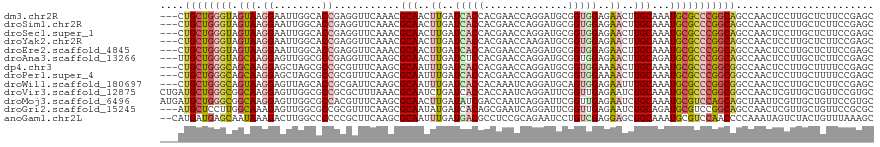
>dm3.chr2R 8808733 116 + 21146708 ---CUGCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))...)).))) ( -45.20, z-score = -2.73, R) >droSim1.chr2R 7268295 116 + 19596830 ---CUGCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))...)).))) ( -45.20, z-score = -2.73, R) >droSec1.super_1 6324357 116 + 14215200 ---CUGCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))...)).))) ( -45.20, z-score = -2.73, R) >droYak2.chr2R 12977873 116 - 21139217 ---CUGCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAAGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))...)).))) ( -45.20, z-score = -3.04, R) >droEre2.scaffold_4845 5573639 116 - 22589142 ---CUGCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))...)).))) ( -45.20, z-score = -2.73, R) >droAna3.scaffold_13266 6276914 116 + 19884421 ---UUGCUGGGUAGCAAGGAGUUGGCGCCGAGGUUCAAGCGCAACUUGAUCUCCACGAACCAGGAUGCGGUGGAGAACUUGCAGAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.((((((((((((((((((.((((((((.....)))))(((((((.............)))))))....((....))))))))).))))))))))))))...)).))) ( -59.82, z-score = -4.93, R) >dp4.chr3 14334204 116 + 19779522 ---CUGCUGGGCAGCAAGGAGCUAGCGCCGCGUUUCAAGCGCAAUUUGAUCACCACGAACCAGGAUGCGGUGGAAAACUUGCAAAUGCGCCCGGCGGCCAACUCCUUGCUUUUCCGAGC ---..(((.((.(((((((((...(.(((((.......(((((.((((.(((((.((........)).)))))........)))))))))...))))))..)))))))))...)).))) ( -42.60, z-score = -0.54, R) >droPer1.super_4 6178996 116 - 7162766 ---CUGCUGGGCAGCAAGGAGCUAGCGCCGCGUUUCAAGCGCAAUUUGAUCACCACGAACCAGGAUGCGGUGGAAAACUUGCAAAUGCGCCCGGCGGCCAACUCCUUGCUUUUCCGAGC ---..(((.((.(((((((((...(.(((((.......(((((.((((.(((((.((........)).)))))........)))))))))...))))))..)))))))))...)).))) ( -42.60, z-score = -0.54, R) >droWil1.scaffold_180697 3078776 116 - 4168966 ---CUGCUGGGCAGUAAGGAGUUAGCACCGCGAUUCAAGCGCAAUUUGAUCACCACAAAUCAGGAUGCAGUGGAGAAUUUGCAAAUGCGCCCGGCGGCCAACUCCUUGCUCUUCCGAGC ---..(((.((.(((((((((((.((.(((.(......(((((.((((((........)))))).(((((........)))))..)))))))))..)).)))))))))))...)).))) ( -40.90, z-score = -1.39, R) >droVir3.scaffold_12875 545823 119 + 20611582 CUGAUGCUGGGCGGCAAGGAGUUGGCGCCGCGCUUUAAACGCAAUCUGAUCACCACCAAUCAGGAUUCGGUUGAGAAUCUGCAAAUGCGCCCGGCGGCCAACUCGUUGCUGUUCCGUGC .....((.(((((((((.(((((((((((((((.......(((..(((((........)))))(((((......))))))))....))))..))).)))))))).))))))))).)).. ( -52.40, z-score = -3.06, R) >droMoj3.scaffold_6496 3544770 119 + 26866924 AUGAUGCUGGGCGGCAAGGAGUUGGCGCCACGUUUCAAGCGCAACUUGAUAUCGACCAAUCAGGAUUCGGUUGAGAAUCUGCAAAUGCGUCCAGCAGCUAAUUCGUUGCUGUUCCGUGC .....((.(((((((((.((((((((((....((((((.((...((((((........))))))...)).))))))....))...(((.....))))))))))).))))))))).)).. ( -45.10, z-score = -2.12, R) >droGri2.scaffold_15245 7906406 116 - 18325388 ---AUGCUCCUUGGCAAAGAGUUGGCGCCGCGUUUCAAGCGCAAUAUGAUCACAGCGAAUCAGGAUUCGGUUGAGAAUCUGCAGAUGCGUCCGGCAGCCAACUCGUUGCUGUUCCGCGC ---..((....((((((.(((((((((((((((.....))))............(((.(((.((((((......))))))...))).)))..))).)))))))).))))))....)).. ( -44.90, z-score = -1.72, R) >anoGam1.chr2L 46268068 117 - 48795086 --CAUGAUGAGCAAUAAAGACUUGGCCCCCCGCUUCAAGCGCAAUUUGAUGACGCCUCCGCAGAAUCCUGUCGAGGAGCUGCAAAUGCGUCCAACCCCAAAUAGUCUACUGUUUAAAGC --.....((((((....((((((((.....(((.....))).........(((((....((((..((((....)))).))))....))))).....)))...)))))..)))))).... ( -29.30, z-score = -1.45, R) >consensus ___CUGCUGGGUAGUAAGGAGUUGGCACCGCGGUUCAAGCGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCCGAGC ....((((((((.(((.((........))...........(((..((..(((((..............)))))..))..)))...)))))))))))....................... (-19.84 = -19.89 + 0.05)
| Location | 8,808,733 – 8,808,849 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Shannon entropy | 0.38026 |
| G+C content | 0.55848 |
| Mean single sequence MFE | -50.15 |
| Consensus MFE | -30.12 |
| Energy contribution | -29.94 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
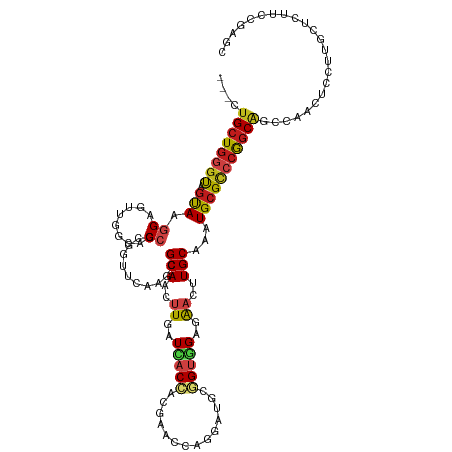
>dm3.chr2R 8808733 116 - 21146708 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).)))))))))))))))............--- ( -53.20, z-score = -4.71, R) >droSim1.chr2R 7268295 116 - 19596830 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).)))))))))))))))............--- ( -53.20, z-score = -4.71, R) >droSec1.super_1 6324357 116 - 14215200 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).)))))))))))))))............--- ( -53.20, z-score = -4.71, R) >droYak2.chr2R 12977873 116 + 21139217 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCUUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).)))))))))))))))............--- ( -53.20, z-score = -4.75, R) >droEre2.scaffold_4845 5573639 116 + 22589142 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).)))))))))))))))............--- ( -53.20, z-score = -4.71, R) >droAna3.scaffold_13266 6276914 116 - 19884421 GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUCUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGAGAUCAAGUUGCGCUUGAACCUCGGCGCCAACUCCUUGCUACCCAGCAA--- (((.((...((((((((((((((.((((((..((..(((((.((((((((.((......))..)))))))...).)))))..))..)).)))))))))))))))))).)).)))..--- ( -63.00, z-score = -6.18, R) >dp4.chr3 14334204 116 - 19779522 GCUCGGAAAAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAGUUUUCCACCGCAUCCUGGUUCGUGGUGAUCAAAUUGCGCUUGAAACGCGGCGCUAGCUCCUUGCUGCCCAGCAG--- (((.((...((((((((((((((.((((.(..((..(((((......((((((..........))))))......)))))..))...).)))))))))))))))))).)).)))..--- ( -57.10, z-score = -4.13, R) >droPer1.super_4 6178996 116 + 7162766 GCUCGGAAAAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAGUUUUCCACCGCAUCCUGGUUCGUGGUGAUCAAAUUGCGCUUGAAACGCGGCGCUAGCUCCUUGCUGCCCAGCAG--- (((.((...((((((((((((((.((((.(..((..(((((......((((((..........))))))......)))))..))...).)))))))))))))))))).)).)))..--- ( -57.10, z-score = -4.13, R) >droWil1.scaffold_180697 3078776 116 + 4168966 GCUCGGAAGAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAAUUCUCCACUGCAUCCUGAUUUGUGGUGAUCAAAUUGCGCUUGAAUCGCGGUGCUAACUCCUUACUGCCCAGCAG--- ((((....))))(((((((((((.((((((..((..(((((......(((..((........))..)))......)))))..))..)).))))))))))))))).((((...))))--- ( -50.20, z-score = -3.83, R) >droVir3.scaffold_12875 545823 119 - 20611582 GCACGGAACAGCAACGAGUUGGCCGCCGGGCGCAUUUGCAGAUUCUCAACCGAAUCCUGAUUGGUGGUGAUCAGAUUGCGUUUAAAGCGCGGCGCCAACUCCUUGCCGCCCAGCAUCAG ((..((....((((.((((((((.((((.((.....(((((((((......)))))((((((......))))))..))))......)).)))))))))))).))))..))..))..... ( -49.20, z-score = -2.82, R) >droMoj3.scaffold_6496 3544770 119 - 26866924 GCACGGAACAGCAACGAAUUAGCUGCUGGACGCAUUUGCAGAUUCUCAACCGAAUCCUGAUUGGUCGAUAUCAAGUUGCGCUUGAAACGUGGCGCCAACUCCUUGCCGCCCAGCAUCAU (((((...(((((.(......).)))))(((.((....(((((((......)))).)))..)))))....(((((.....)))))..)))((((.(((....))).))))..))..... ( -32.50, z-score = -0.09, R) >droGri2.scaffold_15245 7906406 116 + 18325388 GCGCGGAACAGCAACGAGUUGGCUGCCGGACGCAUCUGCAGAUUCUCAACCGAAUCCUGAUUCGCUGUGAUCAUAUUGCGCUUGAAACGCGGCGCCAACUCUUUGCCAAGGAGCAU--- ..((......((((.((((((((.(((((...((..(((((((..(((..(((((....)))))...)))..)).)))))..))...).)))))))))))).))))......))..--- ( -41.10, z-score = -1.45, R) >anoGam1.chr2L 46268068 117 + 48795086 GCUUUAAACAGUAGACUAUUUGGGGUUGGACGCAUUUGCAGCUCCUCGACAGGAUUCUGCGGAGGCGUCAUCAAAUUGCGCUUGAAGCGGGGGGCCAAGUCUUUAUUGCUCAUCAUG-- (((((((...(((....((((((.....(((((.(((((((.((((....))))..))))))).))))).)))))))))..)))))))(..((((.((.......))))))..)...-- ( -35.70, z-score = -0.77, R) >consensus GCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGCUUGAACCGCGGCGCCAACUCCUUACUACCCAGCAG___ ((........))(((((((((((.(((((...((..(((((..((.((((.............)))).)).....)))))..))...).)))))))))))))))............... (-30.12 = -29.94 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:01 2011