| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,806,537 – 8,806,639 |
| Length | 102 |
| Max. P | 0.999028 |

| Location | 8,806,537 – 8,806,639 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Shannon entropy | 0.51171 |
| G+C content | 0.50503 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -25.91 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
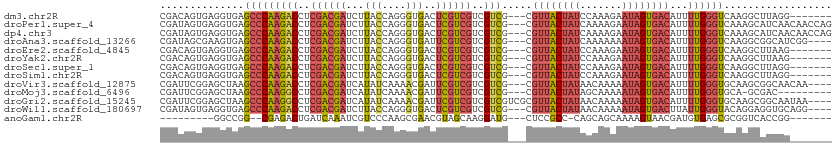
>dm3.chr2R 8806537 102 + 21146708 CGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGGCUUAGG------- ......((((.(..(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))...).))))..------- ( -35.60, z-score = -2.11, R) >droPer1.super_4 6176800 109 - 7162766 CGAUAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCAAAAGAAUAGUGACAUUUUGGGUCAAAGCAUCAACAACCAG .(((.((....(((.((((((((..((((((..((((....)))).))))))..))).---.(((((((((....).))))))))...))))))))..)))))......... ( -37.00, z-score = -3.36, R) >dp4.chr3 14332002 109 + 19779522 CGAUAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCAAAAGAAUAGUGACAUUUUGGGUCAAAGCAUCAACAACCAG .(((.((....(((.((((((((..((((((..((((....)))).))))))..))).---.(((((((((....).))))))))...))))))))..)))))......... ( -37.00, z-score = -3.36, R) >droAna3.scaffold_13266 6274617 105 + 19884421 CGAUAGCGAAGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGAUUCGUCGUCGUCG---CGUUACUAUCAAAAAAAUAGUGACAUUUUGGGUCAAGGCGGCAUCGG---- ((((..((...(((.((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))))...))..)))).---- ( -40.20, z-score = -3.89, R) >droEre2.scaffold_4845 5571414 102 - 22589142 CGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGGCUUAAG------- ......((((.(..(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))...).))))..------- ( -35.60, z-score = -2.46, R) >droYak2.chr2R 12975666 102 - 21139217 CGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGGCUUAAG------- ......((((.(..(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))...).))))..------- ( -35.60, z-score = -2.46, R) >droSec1.super_1 6322168 102 + 14215200 CGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGGCUUAGG------- ......((((.(..(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))...).))))..------- ( -35.60, z-score = -2.11, R) >droSim1.chr2R 7266101 102 + 19596830 CGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGGCUUAGG------- ......((((.(..(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...))))))...).))))..------- ( -35.60, z-score = -2.11, R) >droVir3.scaffold_12875 543408 105 + 20611582 CGAUUCGGAGCUAAGCCCAAGACCUCGACGAUCAUAUCAAAACGAUUCGUCGUCGUCG---CGUUACUAUAACAAAAAUAGUGACAUUUUGGGUGCAAGCGGCAACAA---- .........(((..(((((((((..((((((....(((.....)))))))))..))).---.((((((((.......))))))))...))))))...)))(....)..---- ( -35.10, z-score = -3.86, R) >droMoj3.scaffold_6496 3542404 99 + 26866924 CGAUUCGGAGCUAAGCCCAAGGCCUCGACGAUCAUAUCAAAACGAUUCGUCGUCGUCG---CGUUACUAUAGCAAAAAUAGUGACAUUUUGGGUGCA-GCGAC--------- ......(..(((..(((((((((..(((((((...(((.....)))..)))))))..)---)((((((((.......))))))))..)))))))..)-))..)--------- ( -37.00, z-score = -4.19, R) >droGri2.scaffold_15245 7904104 108 - 18325388 CGAUUCGGAGCUAAGCCCAAGGCCUCGACGAUCAUAUCAAAACGAUUCGUCGUCGUCGUCGCGUUACUAUAACAAAAAUAGUGACAUUUUGGGUGCAAGCGGCAAUAA---- ......(..(((..(((((((((..(((((((...(((.....)))..)))))))..)))..((((((((.......))))))))...))))))...)))..).....---- ( -36.30, z-score = -3.45, R) >droWil1.scaffold_180697 3076574 105 - 4168966 CGAUAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG---CGUUACUAUAACAAAAAUAGUGACUUAUUGGGUACAGGAGGUGCAGG---- ...........((.(((((((((..((((((..((((....)))).))))))..))).---.((((((((.......))))))))...)))))).))...........---- ( -33.70, z-score = -1.55, R) >anoGam1.chr2R 40666115 90 - 62725911 ---------GGCCGG--CGAGACUGAUCAAAUCGUCCCAAGCGAACGUAGCAAGGAUG---CUCCGCC-CAGCAGCAAAAGUAACGAUGUGAGCGCGGUCACCGG------- ---------..((((--...(((((.(((.(((((..(..((.......)).....((---((.(...-..).))))...)..))))).)))...))))).))))------- ( -26.50, z-score = -0.04, R) >consensus CGAUAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCG___CGUUACUAUCAAAAGAAUAGUGACAUUUUGGGUCAAGGCGUCAG_______ ..............(((((((((..((((((...(((....)))..))))))..))).....((((((((.......))))))))...)))))).................. (-25.91 = -26.12 + 0.20)
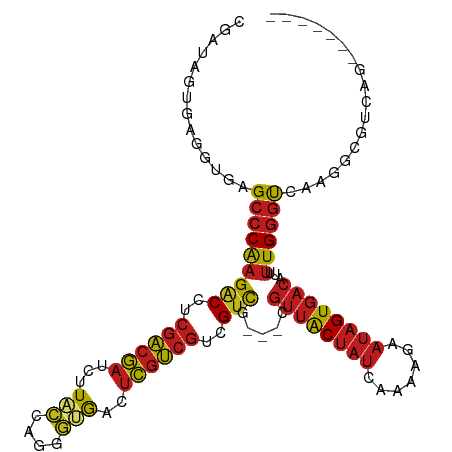

| Location | 8,806,537 – 8,806,639 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Shannon entropy | 0.51171 |
| G+C content | 0.50503 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -16.77 |
| Energy contribution | -16.29 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
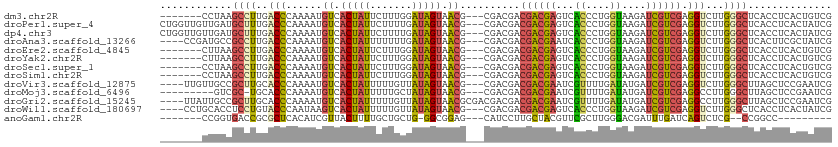
>dm3.chr2R 8806537 102 - 21146708 -------CCUAAGCCUUGACCCAAAAUGUCACUAUUCUUUGGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUGUCG -------....(((((.((((......((.((((((.....)))))).)).---((((((..((.((.....)).))....)))))).))))..)))))............. ( -29.90, z-score = -1.30, R) >droPer1.super_4 6176800 109 + 7162766 CUGGUUGUUGAUGCUUUGACCCAAAAUGUCACUAUUCUUUUGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUAUCG ...(((((((.(((...(((.......)))((((((.....))))))...)---)).)))))))(((((((....))((((((.....)))))).)))))............ ( -29.10, z-score = -0.66, R) >dp4.chr3 14332002 109 - 19779522 CUGGUUGUUGAUGCUUUGACCCAAAAUGUCACUAUUCUUUUGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUAUCG ...(((((((.(((...(((.......)))((((((.....))))))...)---)).)))))))(((((((....))((((((.....)))))).)))))............ ( -29.10, z-score = -0.66, R) >droAna3.scaffold_13266 6274617 105 - 19884421 ----CCGAUGCCGCCUUGACCCAAAAUGUCACUAUUUUUUUGAUAGUAACG---CGACGACGACGAAUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACUUCGCUAUCG ----.(((((.((...((((((((...((.((((((.....)))))).)).---.(((..((((((.((((....))..))))))))..)))))))).)))...)).))))) ( -32.20, z-score = -2.49, R) >droEre2.scaffold_4845 5571414 102 + 22589142 -------CUUAAGCCUUGACCCAAAAUGUCACUAUUCUUUGGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUGUCG -------....(((((.((((......((.((((((.....)))))).)).---((((((..((.((.....)).))....)))))).))))..)))))............. ( -29.90, z-score = -1.35, R) >droYak2.chr2R 12975666 102 + 21139217 -------CUUAAGCCUUGACCCAAAAUGUCACUAUUCUUUGGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUGUCG -------....(((((.((((......((.((((((.....)))))).)).---((((((..((.((.....)).))....)))))).))))..)))))............. ( -29.90, z-score = -1.35, R) >droSec1.super_1 6322168 102 - 14215200 -------CCUAAGCCUUGACCCAAAAUGUCACUAUUCUUUGGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUGUCG -------....(((((.((((......((.((((((.....)))))).)).---((((((..((.((.....)).))....)))))).))))..)))))............. ( -29.90, z-score = -1.30, R) >droSim1.chr2R 7266101 102 - 19596830 -------CCUAAGCCUUGACCCAAAAUGUCACUAUUCUUUGGAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUGUCG -------....(((((.((((......((.((((((.....)))))).)).---((((((..((.((.....)).))....)))))).))))..)))))............. ( -29.90, z-score = -1.30, R) >droVir3.scaffold_12875 543408 105 - 20611582 ----UUGUUGCCGCUUGCACCCAAAAUGUCACUAUUUUUGUUAUAGUAACG---CGACGACGACGAAUCGUUUUGAUAUGAUCGUCGAGGUCUUGGGCUUAGCUCCGAAUCG ----...(((..(((....(((((...((.(((((.......))))).)).---.(((..((((((.((((......))))))))))..))))))))...)))..))).... ( -28.70, z-score = -1.15, R) >droMoj3.scaffold_6496 3542404 99 - 26866924 ---------GUCGC-UGCACCCAAAAUGUCACUAUUUUUGCUAUAGUAACG---CGACGACGACGAAUCGUUUUGAUAUGAUCGUCGAGGCCUUGGGCUUAGCUCCGAAUCG ---------...((-((..(((((...((.(((((.......))))).))(---(..((((((((.(((.....))).)).))))))..)).)))))..))))......... ( -27.90, z-score = -1.31, R) >droGri2.scaffold_15245 7904104 108 + 18325388 ----UUAUUGCCGCUUGCACCCAAAAUGUCACUAUUUUUGUUAUAGUAACGCGACGACGACGACGAAUCGUUUUGAUAUGAUCGUCGAGGCCUUGGGCUUAGCUCCGAAUCG ----...(((..(((....(((((...((((((((.......))))).............((((((.((((......)))))))))).))).)))))...)))..))).... ( -25.80, z-score = -0.25, R) >droWil1.scaffold_180697 3076574 105 + 4168966 ----CCUGCACCUCCUGUACCCAAUAAGUCACUAUUUUUGUUAUAGUAACG---CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUAUCG ----...........((..(((((...((.(((((.......))))).)).---.(((..((((((...((....))....))))))..))))))))..))........... ( -24.50, z-score = -0.27, R) >anoGam1.chr2R 40666115 90 + 62725911 -------CCGGUGACCGCGCUCACAUCGUUACUUUUGCUGCUG-GGCGGAG---CAUCCUUGCUACGUUCGCUUGGGACGAUUUGAUCAGUCUCG--CCGGCC--------- -------(((((((..((..(((.((((((.((......((.(-((((.((---((....)))).)))))))..)))))))).)))...)).)))--))))..--------- ( -31.60, z-score = -0.89, R) >consensus _______CUGAAGCCUUGACCCAAAAUGUCACUAUUCUUUUGAUAGUAACG___CGACGACGACGAGUCACCCUGGUAAGAUCGUCGAGGUCUUGGGCUCACCUCACUAUCG ............((((..(((......((.(((((.......))))).))..........((((((...((....))....)))))).)))...)))).............. (-16.77 = -16.29 + -0.48)
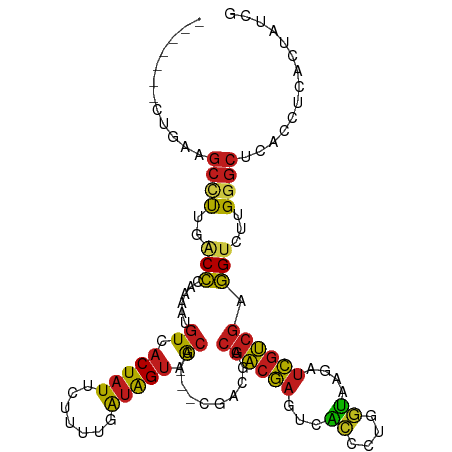
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:59 2011