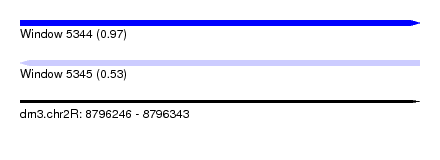
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,796,246 – 8,796,343 |
| Length | 97 |
| Max. P | 0.972743 |

| Location | 8,796,246 – 8,796,343 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.49 |
| Shannon entropy | 0.46690 |
| G+C content | 0.58249 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -18.51 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
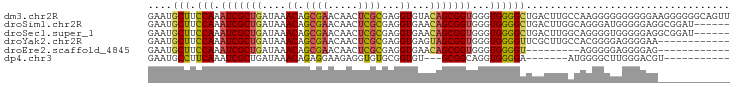
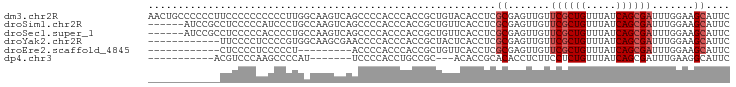
>dm3.chr2R 8796246 97 + 21146708 GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGUACAGCGGUGGGUGGGGCUGACUUGCCAAGGGGGGGGGGAAGGGGGGCAGUU ...(((((((..((((((((((....((((.....))))....))).))))))).((..((.....))..))...............)))))))... ( -35.10, z-score = -2.44, R) >droSim1.chr2R 7256663 91 + 19596830 GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGAACAGCGGUGGGUGGGGCUGACUUGGCAGGGAUGGGGGAGGCGGAU------ ...(((((((...((((((.....)))))).((.(((((.((((...((((..........)))))))).)).))).))..)))))))...------ ( -32.70, z-score = -3.07, R) >droSec1.super_1 6312194 91 + 14215200 GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGAACAGCGGUGGGUGGGGCUGACUUGGCAGGGGUGGGGGAGGCGGAU------ ...(((((((...((((((.....)))))).((.(((((.((((...((((..........)))))))).)).))).))..)))))))...------ ( -32.70, z-score = -2.70, R) >droYak2.chr2R 12964851 85 - 21139217 GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGAGUAGCGGUGGGUGGGGUUCGCUUGCCACGGGGAGGGGAA------------ ...(.(((((...((((((.....))))))...((((((...)))))).(.((..((((.....))))..)).)..))))).)..------------ ( -31.70, z-score = -2.19, R) >droEre2.scaffold_4845 5560793 76 - 22589142 GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGAACAGCGGUGGGUGGGGU---------AGGGGGAGGGGAG------------ ...(.(((((...((((((.....))))))((.(((((((..(....)..).))))))...))---------....))))).)..------------ ( -23.90, z-score = -2.43, R) >dp4.chr3 14819716 76 + 19779522 GAAUGCCUUCAAAUCGCUGAUAAACAGAGGAAGAGGUGUGCGGUGU---GCGGCAGGUGGGGA-------AUGGGGCUUGGGACGU----------- ....((((((..(((((((....(((..(.(.......).)..)))---.)))).)))...))-------...)))).........----------- ( -13.40, z-score = 1.93, R) >consensus GAAUGCUUCCAAAUCGCUGAUAAACAGCGAACAACUCGCGAGGUGAACAGCGGUGGGUGGGGCUGACUUGCCAGGGGGGGGGGAG____________ ....(((.(((.(((((((....((.((((.....))))...))...)))))))...)))))).................................. (-18.51 = -18.68 + 0.17)
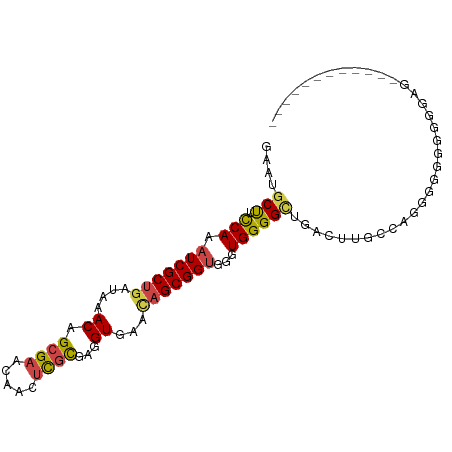
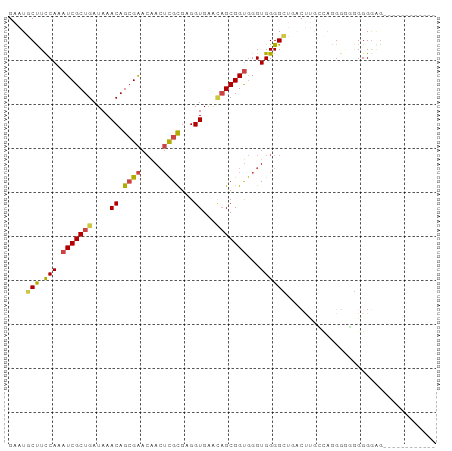
| Location | 8,796,246 – 8,796,343 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.49 |
| Shannon entropy | 0.46690 |
| G+C content | 0.58249 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -9.06 |
| Energy contribution | -9.56 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
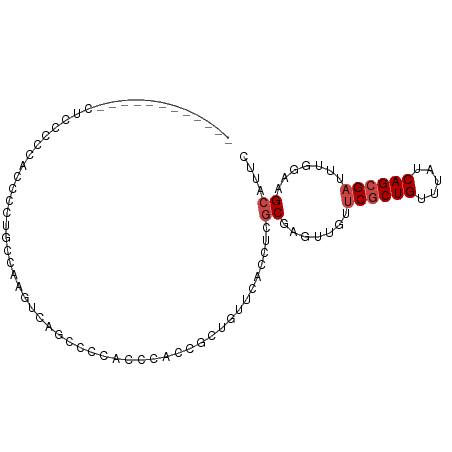
>dm3.chr2R 8796246 97 - 21146708 AACUGCCCCCCUUCCCCCCCCCCUUGGCAAGUCAGCCCCACCCACCGCUGUACACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ..........(((((..........(((......)))........(((((.(((....(((((...))))))))....)))))....)))))..... ( -20.80, z-score = -1.61, R) >droSim1.chr2R 7256663 91 - 19596830 ------AUCCGCCUCCCCCAUCCCUGCCAAGUCAGCCCCACCCACCGCUGUUCACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ------....((.(((...(((.(((..(((.((((...((..(((((..........))).)).))..)))))))..))).)))..))).)).... ( -18.70, z-score = -1.57, R) >droSec1.super_1 6312194 91 - 14215200 ------AUCCGCCUCCCCCACCCCUGCCAAGUCAGCCCCACCCACCGCUGUUCACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ------....((.(((........(((...((((((..........))))...))...)))......((((((.....))))))...))).)).... ( -18.50, z-score = -1.48, R) >droYak2.chr2R 12964851 85 + 21139217 ------------UUCCCCUCCCCGUGGCAAGCGAACCCCACCCACCGCUACUCACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ------------((((.((((..((((..((((............))))..))))...).)))....((((((.....))))))...))))...... ( -19.70, z-score = -1.24, R) >droEre2.scaffold_4845 5560793 76 + 22589142 ------------CUCCCCUCCCCCU---------ACCCCACCCACCGCUGUUCACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ------------.............---------............(((.(((((..(....)..))((((((.....))))))...)))))).... ( -14.10, z-score = -1.68, R) >dp4.chr3 14819716 76 - 19779522 -----------ACGUCCCAAGCCCCAU-------UCCCCACCUGCCGC---ACACCGCACACCUCUUCCUCUGUUUAUCAGCGAUUUGAAGGCAUUC -----------................-------........((((((---.....))..........(.(((.....))).).......))))... ( -9.10, z-score = -0.24, R) >consensus ____________CUCCCCCACCCCUGCCAAGUCAGCCCCACCCACCGCUGUUCACCUCGCGAGUUGUUCGCUGUUUAUCAGCGAUUUGGAAGCAUUC ..........................................................((.......((((((.....)))))).......)).... ( -9.06 = -9.56 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:56 2011