
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,693,989 – 2,694,093 |
| Length | 104 |
| Max. P | 0.981526 |

| Location | 2,693,989 – 2,694,093 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.46273 |
| G+C content | 0.44176 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2693989 104 + 23011544 -GUGUGUGUGUGUGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUU-----UUGCUCUUCUCGUUUUUGCCGCUCCUGGGAUGGUC -.......((((..((((-((....))))))..)))).(((((((.(((.(((((......(.((......-----..)).).......))))).)))..))))))).... ( -27.92, z-score = -1.99, R) >droSim1.chr2L 2653955 110 + 22036055 GGUGUGUGUGUGUGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUUUCGUUUCGCUCCUCUCGUUUUUGCCGCUCCUGGGAGGGCC ((.(((..((((..((((-((....))))))..)))).........(((.(((......))))))............))).))..........(((.(((....)))))). ( -30.40, z-score = -0.95, R) >droSec1.super_5 860818 109 + 5866729 GGUGUAUGUGUGUGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCACUGCGUCGGAAUUUACUGCGCUUCGUUUCGUUUCGCUCCUCUCGUUUUUGCUGCUCCUGG-AGGGCC ((((((.(((((..((((-((....))))))..)))(((((((........)))))))))))))))............((.(((((.((.......))....))-))))). ( -28.40, z-score = -1.10, R) >droAna3.scaffold_12984 140900 100 + 754457 --UGUAUGUGUGCGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUUCGUUUCGC----UCUUUUCGGCUUUCCUCGUCCUUUGGCU---- --.....(((((((((((-((....))))))))).((((((((((....))))))))))..))))........((----(.....(((......)))......))).---- ( -24.80, z-score = -1.48, R) >dp4.chr4_group4 2022691 96 + 6586962 UAUCUGUGUGUGCGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUCCGUUUCGU----UCCUUUUGGCUUUUGUCGUCGU---------- .....(((((((((((((-((....)))))))))))(((((((((....)))))))))...))))..........----.......(((....))).....---------- ( -25.30, z-score = -2.43, R) >droPer1.super_10 1027919 96 + 3432795 UAUCUGUGUGUGCGGUAA-GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUCCGUUUCGU----UCCUUUUGGCUUUUGUCGUCGU---------- .....(((((((((((((-((....)))))))))))(((((((((....)))))))))...))))..........----.......(((....))).....---------- ( -25.30, z-score = -2.43, R) >droWil1.scaffold_180772 1415528 89 + 8906247 UGUUUGUGUGUGUGGAAAAGUUAAUAUUUGCUGCAUAAAUUCCAGGACUUUGGAAUUUUCUGCGCUUGUUCUCGU-----CGUUUUUGUUUUUG----------------- ...........(..((((.(..((((...((.(((.(((((((((....)))))))))..))))).))))..)..-----..))))..).....----------------- ( -15.00, z-score = -0.35, R) >consensus UGUGUGUGUGUGUGGUAA_GUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUUCGUUUCGU____CCCUUUUCGUUUUUGCCGCCCCU_G_______ .......(((((((((((.........))))))).((((((((........))))))))..)))).............................................. (-14.89 = -14.93 + 0.04)

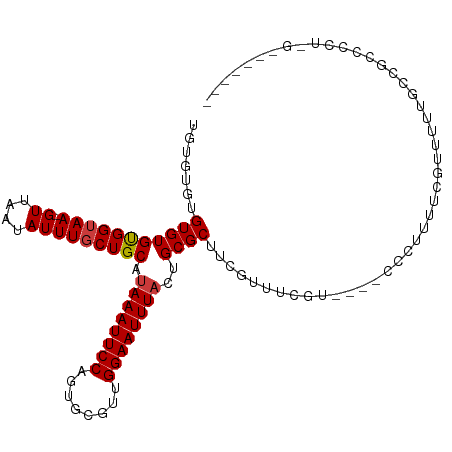
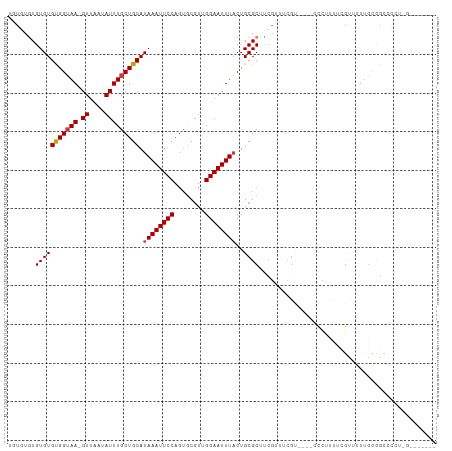
| Location | 2,693,989 – 2,694,093 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.46273 |
| G+C content | 0.44176 |
| Mean single sequence MFE | -18.51 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.03 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2693989 104 - 23011544 GACCAUCCCAGGAGCGGCAAAAACGAGAAGAGCAA-----AACGAAGCGCAGUAAAUUCCGACGCACUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCACACACACACAC- (.((.((....))..)))......(..(((.((..-----......))(((.(((((((((......))))))))))))............)-))..)............- ( -17.40, z-score = -1.45, R) >droSim1.chr2L 2653955 110 - 22036055 GGCCCUCCCAGGAGCGGCAAAAACGAGAGGAGCGAAACGAAACGAAGCGCAGUAAAUUCCGACGCACUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCACACACACACACC .((((((....))).)))..........((..((........))..(((((.(((((((((......)))))))))))).))..........-...))............. ( -25.10, z-score = -2.14, R) >droSec1.super_5 860818 109 - 5866729 GGCCCU-CCAGGAGCAGCAAAAACGAGAGGAGCGAAACGAAACGAAGCGCAGUAAAUUCCGACGCAGUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCACACACAUACACC .(((((-(..(............)..)))).)).............(((((.(((((((((......)))))))))))).))..........-.................. ( -21.70, z-score = -1.38, R) >droAna3.scaffold_12984 140900 100 - 754457 ----AGCCAAAGGACGAGGAAAGCCGAAAAGA----GCGAAACGAAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCGCACACAUACA-- ----.......((..(((....(((.....).----))........(((((.(((((((((......)))))))))))).)).........)-)).))...........-- ( -18.90, z-score = -1.26, R) >dp4.chr4_group4 2022691 96 - 6586962 ----------ACGACGACAAAAGCCAAAAGGA----ACGAAACGGAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCGCACACACAGAUA ----------.((.((.......((....)).----......))..(((((.(((((((((......)))))))))))).))..........-....))............ ( -16.44, z-score = -1.50, R) >droPer1.super_10 1027919 96 - 3432795 ----------ACGACGACAAAAGCCAAAAGGA----ACGAAACGGAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAAC-UUACCGCACACACAGAUA ----------.((.((.......((....)).----......))..(((((.(((((((((......)))))))))))).))..........-....))............ ( -16.44, z-score = -1.50, R) >droWil1.scaffold_180772 1415528 89 - 8906247 -----------------CAAAAACAAAAACG-----ACGAGAACAAGCGCAGAAAAUUCCAAAGUCCUGGAAUUUAUGCAGCAAAUAUUAACUUUUCCACACACACAAACA -----------------..............-----..........(((((..((((((((......)))))))).))).))............................. ( -13.60, z-score = -2.92, R) >consensus _______C_AAGAACGACAAAAACCAAAAGGA____ACGAAACGAAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAAC_UUACCACACACACACACA ..............................................(((((.(((((((((......)))))))))))).))............................. (-14.13 = -14.03 + -0.10)
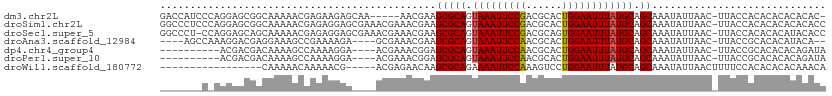
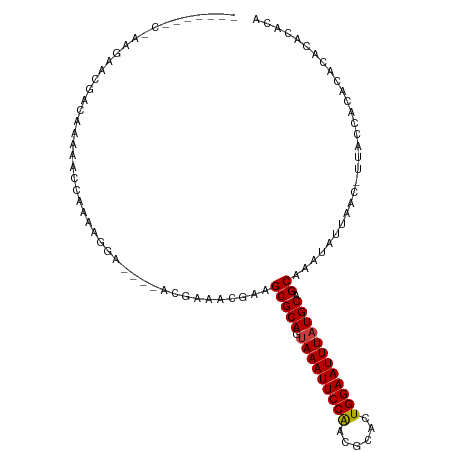
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:03 2011