| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,784,193 – 8,784,285 |
| Length | 92 |
| Max. P | 0.541291 |

| Location | 8,784,193 – 8,784,285 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.96 |
| Shannon entropy | 0.31719 |
| G+C content | 0.55224 |
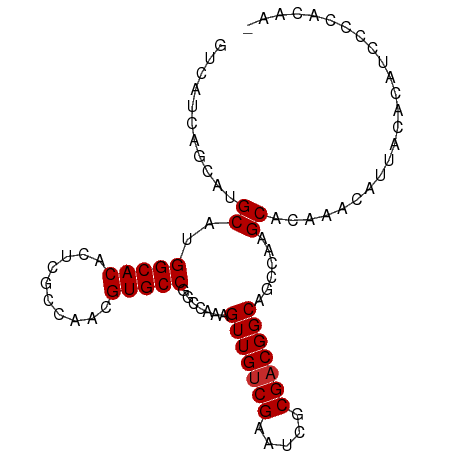
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.61 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
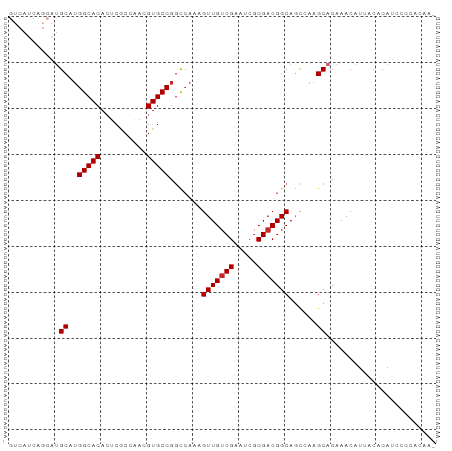
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
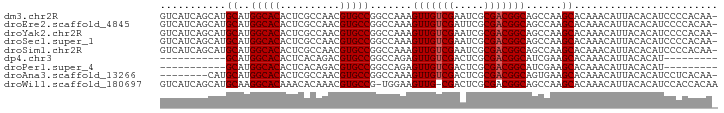
>dm3.chr2R 8784193 92 + 21146708 GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAAUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA- .......(((((..((((......)))).))))).(((....(((((((.....))))))).)))...........................- ( -23.80, z-score = -1.06, R) >droEre2.scaffold_4845 5548002 92 - 22589142 GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAUUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA- .......(((((..((((......)))).))))).(((....(((((((.....))))))).)))...........................- ( -23.80, z-score = -1.03, R) >droYak2.chr2R 12951920 92 - 21139217 GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAAUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA- .......(((((..((((......)))).))))).(((....(((((((.....))))))).)))...........................- ( -23.80, z-score = -1.06, R) >droSec1.super_1 6300335 92 + 14215200 GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAAUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA- .......(((((..((((......)))).))))).(((....(((((((.....))))))).)))...........................- ( -23.80, z-score = -1.06, R) >droSim1.chr2R 7244101 92 + 19596830 GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAAUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA- .......(((((..((((......)))).))))).(((....(((((((.....))))))).)))...........................- ( -23.80, z-score = -1.06, R) >dp4.chr3 14806583 73 + 19779522 -----------GCAUGGCACACUCACAGACGUGCCGGCCAGAGUUGUCGACUCGCGACGGCAUCGAAGCACAAACAUUACACAU--------- -----------((.((((((..........))))))....(((((((((.....))))))).))...))...............--------- ( -20.10, z-score = -1.23, R) >droPer1.super_4 4354696 73 - 7162766 -----------GCAUGGCACACUCACAGACGUGCCGGCCAGAGUUGUCGACUCGCGACGGCAUCGAAGCACAAACAUUACACAU--------- -----------((.((((((..........))))))....(((((((((.....))))))).))...))...............--------- ( -20.10, z-score = -1.23, R) >droAna3.scaffold_13266 6250806 84 + 19884421 --------CAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGACUCGCGACGGCAGUGAAGCACAAACAUUACACAUCCUCACAA- --------..(((.((((((..........))))))..((..(((((((.....)))))))..))..)))......................- ( -20.90, z-score = -0.19, R) >droWil1.scaffold_180697 3039579 91 - 4168966 GUCAUCAGCAUGCAAGGCACAAACACAAACGUGCCG-UGGAAGUUG-CGACUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCACCACAA ...............(((((..........)))))(-((((.((((-((...)))))).((......))..............)))))..... ( -23.10, z-score = -1.29, R) >consensus GUCAUCAGCAUGCAUGGCACACUCGCCAACGUGCCGGCCAAAGUUGUCGAAUCGCGACGGCAGCCAAGCACAAACAUUACACAUCCCCACAA_ ...........((..(((((..........))))).......(((((((.....)))))))......))........................ (-18.50 = -18.61 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:53 2011