| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,783,334 – 8,783,432 |
| Length | 98 |
| Max. P | 0.996149 |

| Location | 8,783,334 – 8,783,432 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.64 |
| Shannon entropy | 0.52068 |
| G+C content | 0.42416 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.14 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
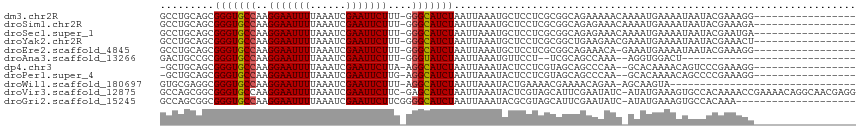
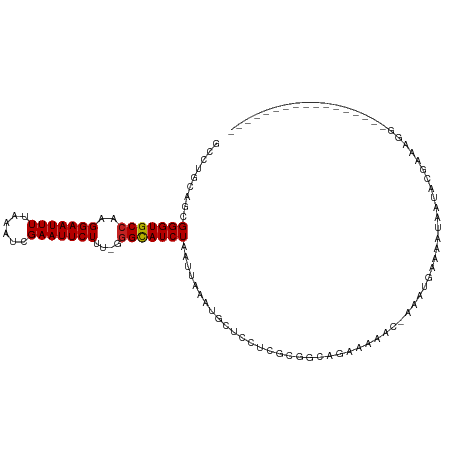
>dm3.chr2R 8783334 98 - 21146708 GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGCAUCUAAUUAAAUGCUCCUCGCGGCAGAAAAACAAAAUGAAAAUAAUACGAAAGG----------------- ..((((.(((((..(((((((((((......)))))))))-))((((........))))..))))).)))).....................(....).----------------- ( -29.90, z-score = -3.45, R) >droSim1.chr2R 7243266 98 - 19596830 GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGCAUCUAAUUAAAUGCUCCUCGCGGCAGAGAAACAAAAUGAAAAUAAUACGAAAGA----------------- ..((((.(((((..(((((((((((......)))))))))-))((((........))))..))))).)))).....................(....).----------------- ( -29.70, z-score = -3.09, R) >droSec1.super_1 6299480 98 - 14215200 GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGCAUCUAAUUAAAUGCUCCUCGCGGCAGAGAAACAAAAUGAAAAUAAUACGAAUGA----------------- ..((((.(((((..(((((((((((......)))))))))-))((((........))))..))))).))))............................----------------- ( -28.50, z-score = -2.66, R) >droYak2.chr2R 12950959 98 + 21139217 GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGCAUCUAAUUAAAUGCUCCUCGCGGCUGAAGAACGAAAUGAAAAUAAUACGAAACU----------------- ..((.((((((((((((((((((((......)))))))))-.))))))).......(((.....))))))).)).........................----------------- ( -25.80, z-score = -1.93, R) >droEre2.scaffold_4845 5547186 97 + 22589142 GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGCAUCUAAUUAAAUGCUCCUCGCGGCAGAAACA-GAAAUGAAAAUAAUACGAAAGG----------------- ..((((.(((((..(((((((((((......)))))))))-))((((........))))..))))).))))...((-....)).........(....).----------------- ( -31.10, z-score = -3.57, R) >droAna3.scaffold_13266 6250188 82 - 19884421 GACUGCCGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-GGGUAUCUAAUUAAAUGUUCCU--UCGCAGCCAAA--AGGUGGACU----------------------------- ..(..((..((.(((.(((((((((((((((.((....))-.)))......))))).))))))--).))).))...--.))..)...----------------------------- ( -22.80, z-score = -1.45, R) >dp4.chr3 14805884 95 - 19779522 -GCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUA-AGGCAUCUAAUUAAAUACUCCUCGUAGCAGCCCAA--GCACAAAACAGUCCCGAAAGG----------------- -(((((...(((((((.((((((((......)))))))).-.))))))).......(((.....))))))))....--.............((....))----------------- ( -27.30, z-score = -2.99, R) >droPer1.super_4 4354021 95 + 7162766 -GCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUG-AGGCAUCUAAUUAAAUACUCCUCGUAGCAGCCCAA--GCACAAAACAGCCCCGAAAGG----------------- -(((((...(((((((.((((((((......)))))))).-.))))))).......(((.....))))))))....--((........)).((....))----------------- ( -29.00, z-score = -2.79, R) >droWil1.scaffold_180697 3037503 83 + 4168966 GUGCGAGGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU-AGGCAUCUAAUUAAAUACUGAAAACGAAAACAGAA-AGCAAGUA------------------------------- .(((...((((((((((((((((((......)))))))))-.))))))).........(((..........)))..-.))..)))------------------------------- ( -21.70, z-score = -3.08, R) >droVir3.scaffold_12875 7312527 114 + 20611582 GCCAGCGGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUC-GAGCAUCUAAUUAAAUACUCGUAGCAUUCGAAUAUC-AUAUGAAAGUGCCACAAAACCGAAAACAGGCAACGAGG (((...)))(((((((.((((((((......)))))))).-).)))))).........(((((.((.((((....((-....))..((........)))))).....)).))))). ( -26.40, z-score = -1.29, R) >droGri2.scaffold_15245 7052907 95 - 18325388 GCCAGCGGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUCGGGGCAUCUAAUUAAAUACGCGUAGCAUUCGAAUAUC-AUAUGAAAGUGCCACAAA-------------------- (((...)))(((((((.((((((((......))))))))...)))))))............((.(((((...(((..-.)))...))))).))...-------------------- ( -23.20, z-score = -0.87, R) >consensus GCCUGCAGCGGGUGCCAAGGAAUUUUAAAUCGAAUUCUUU_GGGCAUCUAAUUAAAUGCUCCUCGCGGCAGAAAAAC_AAAUGAAAAUAAUACGAAAGG_________________ .........(((((((.((((((((......))))))))...)))))))................................................................... (-15.13 = -15.14 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:53 2011