
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,691,361 – 2,691,466 |
| Length | 105 |
| Max. P | 0.983477 |

| Location | 2,691,361 – 2,691,466 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Shannon entropy | 0.21490 |
| G+C content | 0.37867 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.79 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
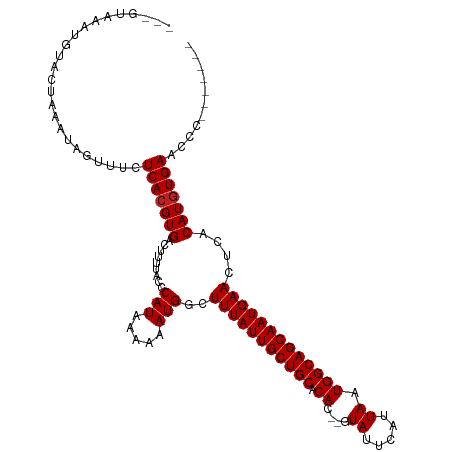
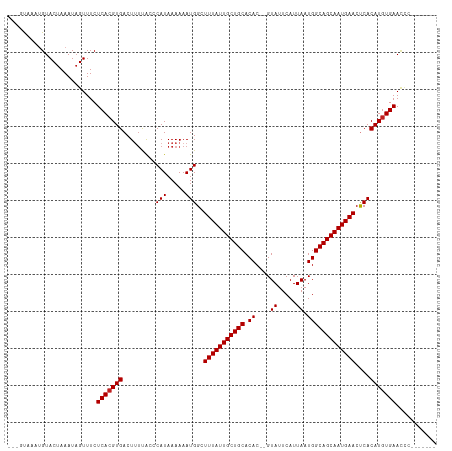
>dm3.chr2L 2691361 105 + 23011544 ---GCAGAUGUACUAAAUAGUUUCUCACGUGACUUUCAACCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCCU------ ---...((...(((....))).))(((((((........((((.....)))).(((((((((((.((.--.((.....)).)))))))))))))....))))))).....------ ( -25.30, z-score = -2.50, R) >droSim1.chr2L 2651276 105 + 22036055 ---GCAGAUGUACUAAAUAGUUUCUCACGUGACUUUCAACCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCCC------ ---...((...(((....))).))(((((((........((((.....)))).(((((((((((.((.--.((.....)).)))))))))))))....))))))).....------ ( -25.30, z-score = -2.49, R) >droSec1.super_5 858188 105 + 5866729 ---GCAGAUGUACUAAAUAGUUUCUCACGUGACUUUCAACCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCCC------ ---...((...(((....))).))(((((((........((((.....)))).(((((((((((.((.--.((.....)).)))))))))))))....))))))).....------ ( -25.30, z-score = -2.49, R) >droYak2.chr2L 2682224 104 + 22324452 ---GCAGAUGUACUAAAUAGUUUCUCACGUGACUUUCAACCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCC------- ---...((...(((....))).))(((((((........((((.....)))).(((((((((((.((.--.((.....)).)))))))))))))....)))))))....------- ( -25.30, z-score = -2.48, R) >droEre2.scaffold_4929 2741319 104 + 26641161 ---GCAGAUGUACUAAAUAGUUUCUCACGUGACUUUCAACCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCC------- ---...((...(((....))).))(((((((........((((.....)))).(((((((((((.((.--.((.....)).)))))))))))))....)))))))....------- ( -25.30, z-score = -2.48, R) >droAna3.scaffold_12984 138214 104 + 754457 ---GUAAAUGUACUAAAUAGUUUCUCACGUGACUUUUGCCCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUAUGAACUC------- ---...............(((((.....((((.....(((..........)))(((((((((((.((.--.((.....)).))))))))))))).))))....))))).------- ( -20.80, z-score = -1.48, R) >dp4.chr4_group4 2019546 105 + 6586962 --AGUAAACGUACUAAAUAGUUUCUCACGUGACUUUUGCCCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACCCACAUGUGAACUC------- --((((....))))..........(((((((..((((........))))(((.(((((((((((.((.--.((.....)).))))))))))))).))))))))))....------- ( -26.40, z-score = -2.77, R) >droPer1.super_10 1024757 105 + 3432795 --AGUAAACGUACUAAAUAGUUUCUCACGUGACUUUUGCCCAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACCCACAUGUGAACUC------- --((((....))))..........(((((((..((((........))))(((.(((((((((((.((.--.((.....)).))))))))))))).))))))))))....------- ( -26.40, z-score = -2.77, R) >droWil1.scaffold_180772 7495713 109 - 8906247 ---GUAACUGUACUAAAUAGUUUCUCACGUGACUUUUAGCCAUAAAAAAUGGCUUUAUUGCUGCACACACGUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAAACUCCC---- ---...............((((((.((.((((......(((((.....)))))(((((((((((.((..............))))))))))))).)))).)).))))))...---- ( -31.24, z-score = -4.44, R) >droVir3.scaffold_12963 8906285 114 - 20206255 GACUUAACUGUACUAAAUAGUUUCUCACGUGACUCUUGCACAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAAUAGAGUGUGG .((((((((((.....))))))..(((((((......((.(((.....)))))(((((((((((.((.--.((.....)).)))))))))))))....)))))))...)))).... ( -29.30, z-score = -2.12, R) >droMoj3.scaffold_6500 12307818 112 - 32352404 GACUUAACUGUACUAAAUAGUUUCUCACGUGACUCCUGCACAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAAUCGAGUGG-- .((((((((((.....))))))..(((((((......((.(((.....)))))(((((((((((.((.--.((.....)).)))))))))))))....)))))))...))))..-- ( -29.30, z-score = -2.75, R) >droGri2.scaffold_15252 11773997 106 - 17193109 GACUUAACUGUACUAAAUUGUUUCUCACGUGACUUUUGCACAUAAAAAAUGGCUUUAUUGCUGCACAC--GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAAUA-------- ........................(((((((......((.(((.....)))))(((((((((((.((.--.((.....)).)))))))))))))....)))))))...-------- ( -24.90, z-score = -2.44, R) >consensus ___GUAAAUGUACUAAAUAGUUUCUCACGUGACUUUUACCCAUAAAAAAUGGCUUUAUUGCUGCACAC__GUAUUCAUUAAUGGCAGCAAUGAACUCACAUGUGAACCC_______ ........................(((((((.........(((.....)))..(((((((((((.((...((....))...)))))))))))))....)))))))........... (-21.71 = -21.79 + 0.08)

| Location | 2,691,361 – 2,691,466 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Shannon entropy | 0.21490 |
| G+C content | 0.37867 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.19 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
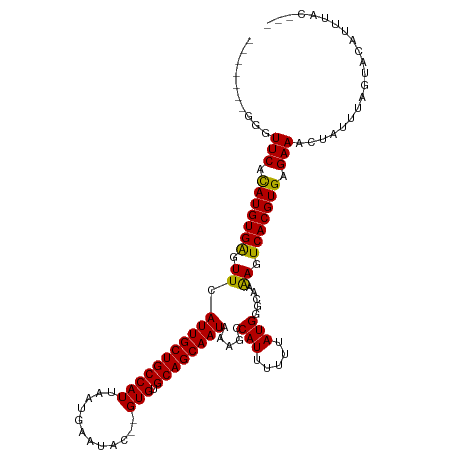
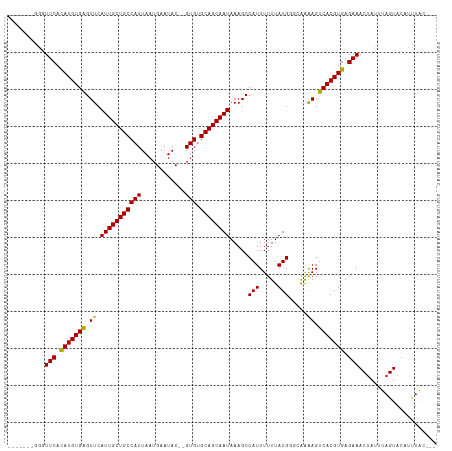
>dm3.chr2L 2691361 105 - 23011544 ------AGGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGUUGAAAGUCACGUGAGAAACUAUUUAGUACAUCUGC--- ------.((.(((.(((((((.((((((((((((((..........--))).))))))))..((((((.....)))))))))..))))))).))).))...............--- ( -31.60, z-score = -2.77, R) >droSim1.chr2L 2651276 105 - 22036055 ------GGGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGUUGAAAGUCACGUGAGAAACUAUUUAGUACAUCUGC--- ------.((.(((.(((((((.((((((((((((((..........--))).))))))))..((((((.....)))))))))..))))))).))).))...............--- ( -31.70, z-score = -2.64, R) >droSec1.super_5 858188 105 - 5866729 ------GGGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGUUGAAAGUCACGUGAGAAACUAUUUAGUACAUCUGC--- ------.((.(((.(((((((.((((((((((((((..........--))).))))))))..((((((.....)))))))))..))))))).))).))...............--- ( -31.70, z-score = -2.64, R) >droYak2.chr2L 2682224 104 - 22324452 -------GGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGUUGAAAGUCACGUGAGAAACUAUUUAGUACAUCUGC--- -------((.(((.(((((((.((((((((((((((..........--))).))))))))..((((((.....)))))))))..))))))).))).))...............--- ( -31.30, z-score = -2.75, R) >droEre2.scaffold_4929 2741319 104 - 26641161 -------GGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGUUGAAAGUCACGUGAGAAACUAUUUAGUACAUCUGC--- -------((.(((.(((((((.((((((((((((((..........--))).))))))))..((((((.....)))))))))..))))))).))).))...............--- ( -31.30, z-score = -2.75, R) >droAna3.scaffold_12984 138214 104 - 754457 -------GAGUUCAUAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGGCAAAAGUCACGUGAGAAACUAUUUAGUACAUUUAC--- -------.(((((.(((((((.((.(((((((((((..........--))).))))))))...(((..........)))..)).))))))).).))))...............--- ( -26.10, z-score = -1.94, R) >dp4.chr4_group4 2019546 105 - 6586962 -------GAGUUCACAUGUGGGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGGCAAAAGUCACGUGAGAAACUAUUUAGUACGUUUACU-- -------.(((..((..((((.((.(((((((((((..........--))).)))))))).)).))))(((((((((((....))).))))))))............))..)))-- ( -28.20, z-score = -1.82, R) >droPer1.super_10 1024757 105 - 3432795 -------GAGUUCACAUGUGGGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGGGCAAAAGUCACGUGAGAAACUAUUUAGUACGUUUACU-- -------.(((..((..((((.((.(((((((((((..........--))).)))))))).)).))))(((((((((((....))).))))))))............))..)))-- ( -28.20, z-score = -1.82, R) >droWil1.scaffold_180772 7495713 109 + 8906247 ----GGGAGUUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUACGUGUGUGCAGCAAUAAAGCCAUUUUUUAUGGCUAAAAGUCACGUGAGAAACUAUUUAGUACAGUUAC--- ----...((((((.(((((((.((.(((((((((((..(((....)))))).))))))))..((((((.....))))))..)).))))))).))))))...............--- ( -37.70, z-score = -4.65, R) >droVir3.scaffold_12963 8906285 114 + 20206255 CCACACUCUAUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGUGCAAGAGUCACGUGAGAAACUAUUUAGUACAGUUAAGUC ..........(((.(((((((.((((((((((((((..........--))).))))))))...(((((.....))).))..)))))))))).)))..................... ( -28.40, z-score = -1.86, R) >droMoj3.scaffold_6500 12307818 112 + 32352404 --CCACUCGAUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGUGCAGGAGUCACGUGAGAAACUAUUUAGUACAGUUAAGUC --..(((..((((.(((((((.((((((((((((((..........--))).))))))))...(((((.....))).)).))).))))))).)).(((....)))...))..))). ( -31.10, z-score = -2.47, R) >droGri2.scaffold_15252 11773997 106 + 17193109 --------UAUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC--GUGUGCAGCAAUAAAGCCAUUUUUUAUGUGCAAAAGUCACGUGAGAAACAAUUUAGUACAGUUAAGUC --------..(((.(((((((.((.(((((((((((..........--))).))))))))...(((((.....))).))..)).))))))).)))..................... ( -27.10, z-score = -2.31, R) >consensus _______GGGUUCACAUGUGAGUUCAUUGCUGCCAUUAAUGAAUAC__GUGUGCAGCAAUAAAGCCAUUUUUUAUGGGCAAAAGUCACGUGAGAAACUAUUUAGUACAUUUAC___ ..........(((.(((((((.((.(((((((((((............))).)))))))).....(((.....))).....)).))))))).)))..................... (-23.54 = -23.19 + -0.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:02 2011