| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,691,606 – 8,691,666 |
| Length | 60 |
| Max. P | 0.979029 |

| Location | 8,691,606 – 8,691,666 |
|---|---|
| Length | 60 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.62455 |
| G+C content | 0.36219 |
| Mean single sequence MFE | -11.81 |
| Consensus MFE | -7.99 |
| Energy contribution | -8.09 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979029 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 8691606 60 - 21146708 GUGUGCGUGGCAUAUGUAAAUUUCCAGCUAAUUAAAAUUAGCAUAUAUUUCUCUCCGCAU----------------- ..((((((((...((....))..)))((((((....)))))).............)))))----------------- ( -11.20, z-score = -1.37, R) >droSim1.chr2R 7163040 71 - 19596830 GUGUGCGUGGCAUAUGUAAAUUUCCAGCUAAUUAAAAUUAGCAUAUAUUU-CCCCCAAUCCCCACUCCACAC----- (((((.((((....((........))((((((....))))))........-..........))))..)))))----- ( -13.90, z-score = -3.39, R) >droSec1.super_1 6223826 72 - 14215200 GUGUGCAUGGCAUAUGUAAAUUUCCAGCUAAUUAAAAUUAGCAUAUAUUUCCCCCCAAUUCCCUCUACACAU----- (((((..(((...((....))..)))((((((....)))))).......................)))))..----- ( -10.70, z-score = -1.93, R) >droYak2.chr2R 12872763 64 + 21139217 GUGUGCGUGGCAUAUGUAAAUUUGGAGCUAAUUAAAAUUAGCAUAUA-UUCCCCUUGCCCCAUAU------------ (((((.(.((((...(..(((.((..((((((....)))))).)).)-))..)..))))))))))------------ ( -13.70, z-score = -1.05, R) >droEre2.scaffold_4845 5463540 69 + 22589142 GUGUGCGUGGCAUAUGUAAAUUUGGAGCUAAUUAAAAUUAGCAUAUAUUUCCCCUUUUCACAUGUAUAU-------- (((((((((((....)).........((((((....))))))..................)))))))))-------- ( -13.60, z-score = -0.96, R) >droAna3.scaffold_13266 6161214 69 - 19884421 GUGUGUGUGGCAUAUGUAAAUUUCAAGCUAAUUAAAAUUAGCAUAUAUUUUCCCCUUUUGCGAGGACAU-------- (((((.....)))))...........((((((....))))))...........((((....))))....-------- ( -10.70, z-score = 0.11, R) >dp4.chr3 14728671 68 - 19779522 GUGUGCUUGGCAUAUGUAGAUUUCCUGCUAAUUAAAAUUAGCAUACAUAUGAUUCCCUCUCCCCGGAU--------- ..........((((((((.......(((((((....)))))))))))))))..(((........))).--------- ( -13.81, z-score = -1.31, R) >droPer1.super_4 4276068 68 + 7162766 GUGUGCUUGGCAUAUGUAGAUUUCCUGCUAAUUAAAAUUAGCAUACAUAUGAUUCCCUCUCCCCGGAU--------- ..........((((((((.......(((((((....)))))))))))))))..(((........))).--------- ( -13.81, z-score = -1.31, R) >droWil1.scaffold_180708 12364158 74 - 12563649 GUGCGCGUGGCAUAUGUAAAUUUCCAACUAAUUAAAAUUAGCAUA---UUCCUCUUGCAACUACUCCAGCUCCACCU .((((.(.((.((((((.(((((...........))))).)))))---).)).).)))).................. ( -12.60, z-score = -1.92, R) >droVir3.scaffold_12875 15752912 58 - 20611582 GUGUGCGUAGCAUAUGUAAAUUUUUGGCUAAUUAAAAUUAGCAUA---UUCCCUUUGGCUU---------------- ((((((...))))))...........((((((....))))))...---.............---------------- ( -9.00, z-score = 0.30, R) >droMoj3.scaffold_6496 18837164 58 + 26866924 GUGUGCGUAGCAUAUGUAAAUUUUUGGCUAAUUAAAAUUAGCAUA---UUCCCUUUAGCAU---------------- ..((((..((.((((((.((((((.........)))))).)))))---)...))...))))---------------- ( -9.60, z-score = -0.21, R) >droGri2.scaffold_15245 12569128 58 - 18325388 GAGUGCGUAGCACAUGUAAAUUUCUGGCUAAUUAAAAUUAGCAUA---UUCCCUUUAGCGU---------------- ..((((...)))).............((((((....))))))...---.............---------------- ( -9.10, z-score = -0.03, R) >consensus GUGUGCGUGGCAUAUGUAAAUUUCCAGCUAAUUAAAAUUAGCAUAUAUUUCCCCCUAUCUC________________ ((((((...))))))...........((((((....))))))................................... ( -7.99 = -8.09 + 0.10)

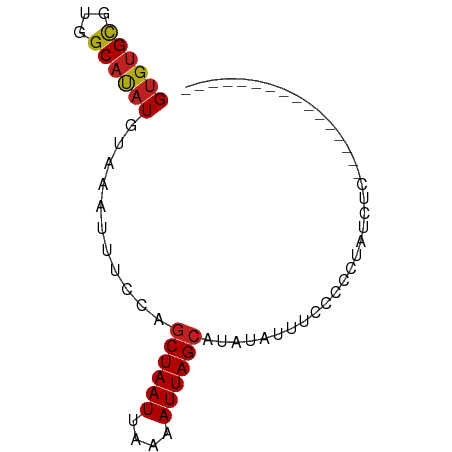
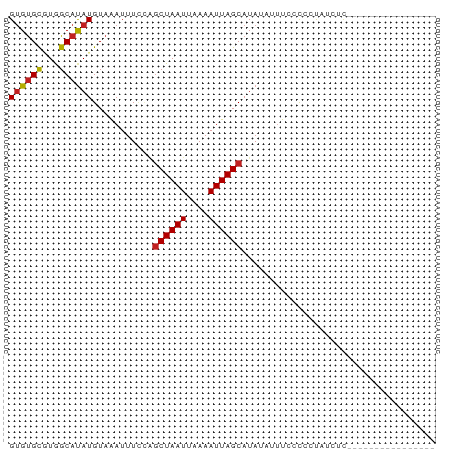
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:45 2011