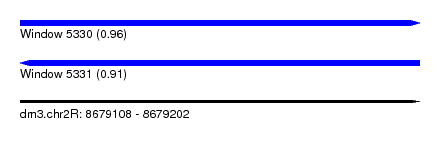
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,679,108 – 8,679,202 |
| Length | 94 |
| Max. P | 0.956257 |

| Location | 8,679,108 – 8,679,202 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 56.23 |
| Shannon entropy | 0.72961 |
| G+C content | 0.48059 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -12.53 |
| Energy contribution | -11.47 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
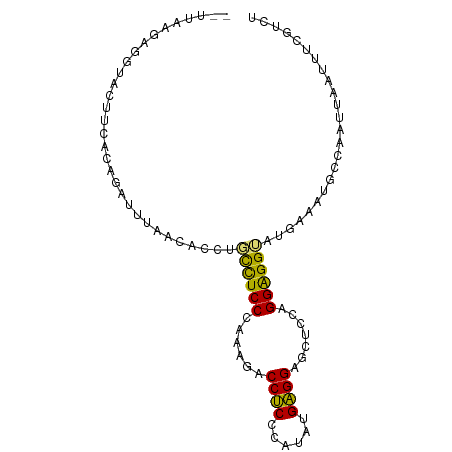
>dm3.chr2R 8679108 94 + 21146708 CUUUAAAUGGUAUUUCAUUGACUUAACUCCAGCUUCCCAAAUACCUCCGAAGUGAGGUCCUGCCGGGGGAAUAAAAUGCCAAUCAAUCCUGUCC .......((((((((..(((.........))).(((((....(((((......)))))((....)))))))..))))))))............. ( -18.70, z-score = 0.14, R) >droYak2.chr2h 381189 86 + 894407 -------AGGUGCUUUGCAGUUUUCACACCUGCCUCCCAUAGUCCGCCCAUGUGCGGAGCCCGAGGGGGUGUGAA-CGCAAAUUCGCAUCCCCU -------.((((((((((.((..((((((((.((((......(((((......)))))....)))))))))))))-))))))...))))).... ( -34.60, z-score = -2.65, R) >droAna3.scaffold_12943 4861664 92 - 5039921 --UUAGGACUGAUUUAACAGAUCGAACAAGCCUUUCCCACGCUCCUCCCAUAUGAGGCGCUGCCGGAGGGUUGAAUUUCCAUAUAAUUGCGUCA --...(((.(((((.....)))))..(((.(((((..((.((.((((......)))).))))..))))).)))....))).............. ( -22.70, z-score = -0.62, R) >triCas2.singleUn_1341 5284 94 + 21755 CGUUAAGUGAAACUUCAUAGAUUUAAUUCCUGCCUCCCAAAGACCUCCCAUAUGAGGGGCUCUAGGAGGUAUGAAAUGCCAACUAAUUUUUUCU .(((..((((....))))........(((.(((((((...(((.(((((....).)))).))).))))))).))).....)))........... ( -21.60, z-score = -0.85, R) >consensus __UUAAGAGGUACUUCACAGAUUUAACACCUGCCUCCCAAAGACCUCCCAUAUGAGGAGCUCCAGGAGGUAUGAAAUGCCAAUUAAUUUCGUCU ...............................((((((......((((......)))).......))))))........................ (-12.53 = -11.47 + -1.06)

| Location | 8,679,108 – 8,679,202 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 56.23 |
| Shannon entropy | 0.72961 |
| G+C content | 0.48059 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -12.46 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8679108 94 - 21146708 GGACAGGAUUGAUUGGCAUUUUAUUCCCCCGGCAGGACCUCACUUCGGAGGUAUUUGGGAAGCUGGAGUUAAGUCAAUGAAAUACCAUUUAAAG .....(((((.((((((.....((((((((((....(((((......)))))..))))).....)))))...))))))..))).))........ ( -23.20, z-score = -0.08, R) >droYak2.chr2h 381189 86 - 894407 AGGGGAUGCGAAUUUGCG-UUCACACCCCCUCGGGCUCCGCACAUGGGCGGACUAUGGGAGGCAGGUGUGAAAACUGCAAAGCACCU------- ..(((.(((...((((((-((((((((.((((.((.(((((......)))))))....))))..))))))))...))))))))))))------- ( -35.90, z-score = -2.05, R) >droAna3.scaffold_12943 4861664 92 + 5039921 UGACGCAAUUAUAUGGAAAUUCAACCCUCCGGCAGCGCCUCAUAUGGGAGGAGCGUGGGAAAGGCUUGUUCGAUCUGUUAAAUCAGUCCUAA-- .(((........(((((....(((.(((((.((.((.((((......)))).)))).)))..)).))).....))))).......)))....-- ( -22.76, z-score = 0.38, R) >triCas2.singleUn_1341 5284 94 - 21755 AGAAAAAAUUAGUUGGCAUUUCAUACCUCCUAGAGCCCCUCAUAUGGGAGGUCUUUGGGAGGCAGGAAUUAAAUCUAUGAAGUUUCACUUAACG ........((((.(((.(((((((.((((((((((((.(((....))).)).))))))))))..(((......)))))))))).))).)))).. ( -29.00, z-score = -2.50, R) >consensus AGACAAAAUUAAUUGGCAUUUCAUACCCCCGGCAGCACCUCACAUGGGAGGACUUUGGGAAGCAGGAGUUAAAUCUAUGAAAUACCACUUAA__ .........................((((((((((.(((((......))))))))))))))).(((((((..........)))))))....... (-12.46 = -14.40 + 1.94)

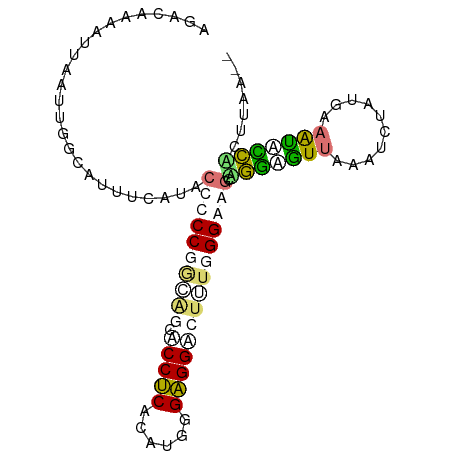
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:44 2011