
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,647,946 – 8,648,044 |
| Length | 98 |
| Max. P | 0.530119 |

| Location | 8,647,946 – 8,648,044 |
|---|---|
| Length | 98 |
| Sequences | 14 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.30 |
| Shannon entropy | 0.79149 |
| G+C content | 0.53782 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -11.39 |
| Energy contribution | -10.84 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
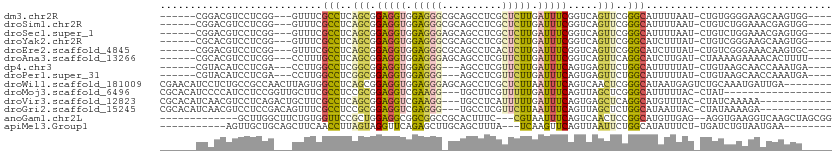
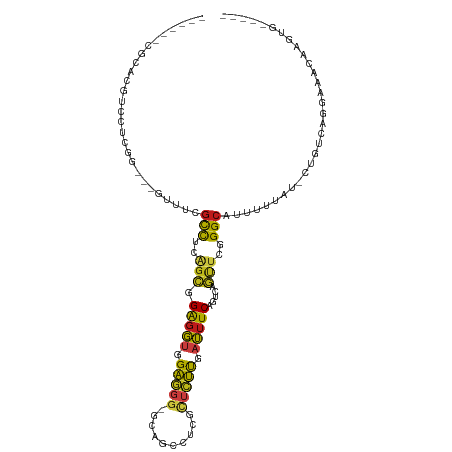
>dm3.chr2R 8647946 98 + 21146708 ------CGGACGUCCUCGG---GUUUCGCCUCAGCGGAGGUGGAGGGCGCAGCCUCGCUCUUGAUUUCGGUCAGUUCGGGCAUUUUAAU-CUGUGGGGAAGCAAGUGG---- ------...((((((((.(---.((((((....)))))).).))))))((..((((((....((((...(((......))).....)))-).))))))..))..))..---- ( -36.00, z-score = -0.64, R) >droSim1.chr2R 7128613 98 + 19596830 ------CGGACGUCCUCGG---GUUUCGCCUCAGCGGAGGUGGAGGGCGCAGCCUCGCUCUUGAUUUCGGUCAGUUCGGGCAUUUUAAU-CUGUCUGGAAACGAGUGG---- ------.((.(((((((.(---.((((((....)))))).).)))))))...)).((((((((((....)))))((((((((.......-.))))))))...))))).---- ( -39.40, z-score = -1.72, R) >droSec1.super_1 6185940 98 + 14215200 ------CGGACGUCCUCGG---GUUUCGCCUCAGCGGAGGUGGAGGGAGCAGCCUCGCUCUUGAUUUCGGUCAGUUCGGGCAUUUUAAU-CUGUCUGGAAACGAGUGG---- ------.((.(..((((.(---.((((((....)))))).).))))..)...)).((((((((((....)))))((((((((.......-.))))))))...))))).---- ( -34.30, z-score = -0.25, R) >droYak2.chr2R 12837200 98 - 21139217 ------CGCACGUCCUCGG---GUUUCGCCUCAGCGGAGGUGGAGGGCGCAGCCUCGCUCUUGAUUUCGGUCAGUUCGGGCAUCUUUAU-CUGUCGGGAAGCAAGUGG---- ------..(((((((((.(---.((((((....)))))).).))))))((.(((.((..((.(((....)))))..))))).((((...-.....)))).))..))).---- ( -35.10, z-score = -0.31, R) >droEre2.scaffold_4845 5428371 98 - 22589142 ------CGGACGUCCUCGG---GUUUCGCCUCAGCGGAGGUGGAGGGCGCAGCCUCACUCUUGAUUUCGGUCAGUUCGGGCAUCUUUAU-CUGUCGGGAAACAAGUGC---- ------.((.(((((((.(---.((((((....)))))).).)))))))...))...(.((((.((((.(.(((...((....))....-))).).)))).)))).).---- ( -33.60, z-score = -0.35, R) >droAna3.scaffold_13266 11391033 98 - 19884421 ------CGCACGUCCUCGG---CCUUUGCCUCAGCGGAGGUGGAGGGAGCAGCCUCGUUCUUGAUUUCGGUCAGUUCAGGCAUCUUGAU-CUAAAAGAAAACACUUUU---- ------.((...(((((.(---(((((((....)))))))).))))).)).((((.(..((.(((....)))))..))))).((((...-....))))..........---- ( -33.50, z-score = -1.53, R) >dp4.chr3 2488050 95 - 19779522 ------CGUACAUCCUCGA---CCUUGGCCUCGGCGGAGGUGGAGGG---AGCCUCGUUCUUGAUUUCAGUGAGUUCUGGCAUUUUUAU-CUGUAAGCAACCAAAUGA---- ------..(((((((((.(---((((.((....)).))))).)))))---.(((...((((((....))).)))....)))........-.)))).............---- ( -29.60, z-score = -0.81, R) >droPer1.super_31 499340 95 + 935084 ------CGUACAUCCUCGA---CCUUGGCCUCGGCGGAGGUGGAGGG---AGCCUCGUUCUUGAUUUCAGUGAGUUCUGGCAUUUUUAU-CUGUAAGCAACCAAAUGA---- ------..(((((((((.(---((((.((....)).))))).)))))---.(((...((((((....))).)))....)))........-.)))).............---- ( -29.60, z-score = -0.81, R) >droWil1.scaffold_181009 1699097 104 - 3585778 CGAACAUCCUCUGCCGCCAACUUAGUGGCCUCAGCGGAGGUGGAGGGAGCAGCCUCGCUCUUAAUUUCAGUCAACUCGGGCAUAAUGAGUCUGCAAAUGAUUGA-------- ....((.(((((((.((((......))))....)))))))))..((((((......))))))....(((((((...(((((.......)))))....)))))))-------- ( -34.90, z-score = -0.98, R) >droMoj3.scaffold_6496 258237 90 + 26866924 CGCACAUCCCCAUCCUCCGGUUGCUUCGCCUCCGCGGAGGUCGAAGG---UGCUUCGUUUUUGAUUUCAGUUAGCUCGGGCAUUUUUAC-CUAU------------------ ..................(((..(((((((((....)))).)))))(---(((((.(((.(((....)))..)))..))))))....))-)...------------------ ( -22.60, z-score = -0.06, R) >droVir3.scaffold_12823 444656 96 + 2474545 CGCACAUCAACGUCCUCAGACUGCUUCGCCUCAGCGGAGGUCGAAGG---UGCCUCAUUUUUGAUUUCAGUGAGCUCAGGCAUGUUUAC-CUAUCAAAAA------------ .((((......(((....)))..(((((((((....)))).))))))---)))....((((((((....((((((........))))))-..))))))))------------ ( -29.90, z-score = -2.30, R) >droGri2.scaffold_15245 12109227 96 - 18325388 CGCACAUCAACGUCCUCCGACAGUUUCGCCUCCGCGGAGGUCGAGGG---UGCCUCGUUCUUAAUUUCAGUUAGCUCUGGCAUAAUUAC-CUAUAAAAGA------------ .((.((((..((.((((((..((......))...)))))).))..))---))...............(((......)))))........-..........------------ ( -22.50, z-score = -0.20, R) >anoGam1.chr2L 11325413 94 - 48795086 -------------GCUUGGCUUCUGUGGUUCCGCUGGAGGCGGCGGCCGCACUUUC---CGUAAUUUCAGUCAACUCCGGCAUGUUGAG--AGGUGAAGGUCAAGCUAGCGG -------------(((((((((((((((((((((.....)))).))))))).....---....(((((..(((((........))))))--))))))).)))))))...... ( -33.40, z-score = -0.27, R) >apiMel3.Group1 15822848 89 + 25854376 -----------AGUUGCUGCAGCUUCAACCUUAGUAGGUUCAGAGCUUGCAGCUUUA---UCAAGUUCAGUUAAUUCUGGCAUAUUUCU-UGAUCUGUAAUGAA-------- -----------....((((((((((.(((((....)))))..)))).))))))...(---(((((.((((......)))).......))-))))..........-------- ( -24.30, z-score = -1.55, R) >consensus ______CGCACGUCCUCGG___GUUUCGCCUCAGCGGAGGUGGAGGG_GCAGCCUCGCUCUUGAUUUCAGUCAGUUCGGGCAUUUUUAU_CUGUCAGGAAACAAGUG_____ ...........................(((..(((.(((((.(((((..........))))).))))).....)))..)))............................... (-11.39 = -10.84 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:38 2011