| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,641,249 – 8,641,360 |
| Length | 111 |
| Max. P | 0.500000 |

| Location | 8,641,249 – 8,641,360 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.28 |
| Shannon entropy | 0.68985 |
| G+C content | 0.50151 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -13.46 |
| Energy contribution | -12.79 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 8641249 111 - 21146708 GUCCACAAUAAACCGAGGCAUCAUUCUUUCCAUUCGCAGGCAAGUGCAUACCUGUUGUGCGUGGCAUCGGUGUCUACCAGGAUGCCAUCAACCUGUGCAUCGAGAAGGCCG ................(((....(((((...((.((((((.....(((((.....)))))(((((((((((....)))..))))))))...)))))).)).))))).))). ( -34.30, z-score = -1.01, R) >droMoj3.scaffold_6496 24665488 105 + 26866924 -----AUAUAAGGUACUAUAAAUGCAUAAUUA-UUAUAGGUAAAUGCAUACCUGUGGUGCGUGGCAAUGGCGUCUACCAGGAGGCCAUUAAUCUGUGCAUCGAAAAGUGCG -----.......(((((...............-..(((((((......)))))))((((((..(.((((((.(((....))).))))))...)..))))))....))))). ( -34.40, z-score = -2.78, R) >droVir3.scaffold_10324 897668 105 + 1288806 -----AUAUUUGAUAC-AUAAAUGUGUGUGUAUUUGCAGGCAAAUGCAUACCGGUUGUGCGGGGCCAUGGCGUUUAUCAGGAGGCAAUCAAUCUGUGCAUCGAGAAGUGCG -----......(.(((-........(((((((((((....)))))))))))((((.(..((((......((.(((....))).))......))))..)))))....)))). ( -28.50, z-score = 0.31, R) >droWil1.scaffold_180700 6128314 93 + 6630534 ------------------UAAUUCUCUUUGAUUGCUUAGGCAAAUGUAUCCCUGUCGUGCGAGGUCUUGGUGUCUAUCAGGAAGCAAUUAAUUUGUGUAUAGAGAAAUGCG ------------------...(((((((((((((((..((((..........))))........(((((((....)))))))))))))))).........))))))..... ( -20.00, z-score = -0.39, R) >droPer1.super_4 766771 110 + 7162766 -UCCACUGAUGAUCCUAAUCGAUCCGAUCUUCACUGCAGGCAAGUGUAUUCCUGUUGUGCGCGGCUCCGGUGUCUACCAGGAGGCCAUCAACCUGUGCAUCGAAAAGUGCG -.....(((.((((...........)))).)))..(((((...((((((.......))))))(((((((((....))).))).))).....)))))((((......)))). ( -30.80, z-score = -0.20, R) >dp4.chr3 8702485 110 + 19779522 -UCCACUGAUGAUCCGAACCGAUCCGAUCUUCUCUGCAGGCAAGUGUAUUCCUGUUGUGCGCGGCUCCGGUGUCUACCAGGAGGCCAUCAACCUGUGCAUCGAAAAGUGCG -......((.((((.((.....)).)))).))...(((((...((((((.......))))))(((((((((....))).))).))).....)))))((((......)))). ( -32.20, z-score = -0.34, R) >droAna3.scaffold_13266 11383983 91 + 19884421 -------------------AUCACUGUUUUCCUCC-CAGGUAAGUGCAUACCUGUUGUGAGAGGCAACGGUGUCUACCAAGAAGCCAUUAACCUGUGUAUCGAGAAGGCUG -------------------..(((((((..(((((-((((((......))))))....).)))).)))))))..........((((....................)))). ( -21.75, z-score = -0.03, R) >droEre2.scaffold_4845 5421744 94 + 22589142 ---------------ACCCACAACU--UUCCAUUUGCAGGCAAGUGCAUACCUGUGGUGCGUGGCAUCGGUGUCUACCAGAAUGCCAUUAACCUGUGCAUCGAGAAGGCCG ---------------..(((((...--...((((((....))))))......))))).....(((.(((((((....(((............))).)))))))....))). ( -25.00, z-score = 0.54, R) >droYak2.chr2R 12830537 107 + 21139217 --ACCCAAGGAAAUCAGAGCCAAUU--UUUCAUUCGCAGGCAAGUGCAUACCUGUGGUGCGUGGCAUCGGUGUCUACCAGGAUGCCAUCAACCUAUGCAUCGAGAAGGCCG --..............(.(((...(--((((....(((((..........)))))(((((((((....(((((((....)))))))......)))))))))))))))))). ( -30.90, z-score = -0.28, R) >droSec1.super_1 6179249 111 - 14215200 GGCCACAAUAAAACGAGCCAUCAUUCUUUCCAUCCGCAGGCAAGUGCAUACCAGUUGUGCGUGGCAUCGGCGUCUACCAGGAUGCCAUCAACCUGUGCAUCGAGAAGGCCG ((((..........((....))....((((.((.((((((.....(((((.....)))))((((((((((......))..))))))))...)))))).)).)))).)))). ( -36.30, z-score = -1.64, R) >droSim1.chr2R 7121925 109 - 19596830 --CCACAAUAAACCGAGCCAUCAUUCUUUCCAUCCGCAGGCAAGUGCAUACCAGUUGUGCGUGGCAUCGGUGUCUACCAGGAUGCCAUCAACCUGUGCAUCGAGAAGGCCG --...........((.(((....(((((...((.((((((.....(((((.....)))))(((((((((((....)))..))))))))...)))))).)).)))))))))) ( -32.30, z-score = -1.21, R) >consensus ___C_CAAUAAA_C_AGACAUAACUCUUUUCAUCCGCAGGCAAGUGCAUACCUGUUGUGCGUGGCAUCGGUGUCUACCAGGAUGCCAUCAACCUGUGCAUCGAGAAGGCCG ..................................(((((((...(((((.......)))))..))...(((.(((....))).)))......))))).............. (-13.46 = -12.79 + -0.67)

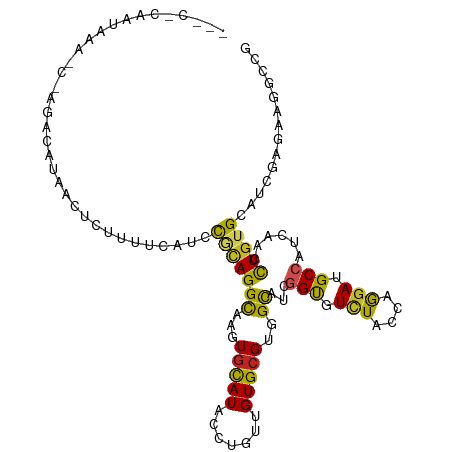

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:37 2011