| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,618,917 – 8,619,073 |
| Length | 156 |
| Max. P | 0.619846 |

| Location | 8,618,917 – 8,619,073 |
|---|---|
| Length | 156 |
| Sequences | 7 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 66.04 |
| Shannon entropy | 0.61803 |
| G+C content | 0.54072 |
| Mean single sequence MFE | -48.33 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
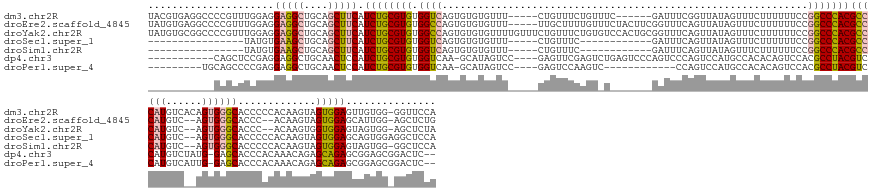
>dm3.chr2R 8618917 156 + 21146708 UACGUGAGGCCCCGUUUGGAGGAGGCUGCAGCUUCAUCUGCGUGUGGUCAGUGUGUGUUU-----CUGUUUCUGUUUC------GAUUUCGGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUCACAGUGGGCACCCCCACAAGUAGUGGAGUUGUGG-GGUUCCA ...(((.((((((....)).((((((..((((.......)).))..)))...........-----((((..(((....------.....)))..)))).........))))))))))((((((......))))))..((((((((.(.....).))))))-))..... ( -54.10, z-score = -1.31, R) >droEre2.scaffold_4845 5399439 158 - 22589142 UAUGUGAGGCCCCGUUUGGAGGAGGCUGCAGCUUCAUCUGCGUGUGGCCAGUGUGUGUUU-----UUGCUUUUGUUUCUACUUCGGUUUCAGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUC--AGUGGGCACCC--ACAAGUAGUGGAGCAUUGG-AGCUCUG ....((.((((((.......)).)))).))((((((..((((((.((((((((.......-----.)))).((((..((((....))...))..))))............)))))))((((((...--.))))))..((--((.....)))).))).)))-))).... ( -51.20, z-score = -0.68, R) >droYak2.chr2R 12807487 163 - 21139217 UAUGUGCGGCCCCGUUUGGAGGAGGCUGCAGCUUCAUCUGCGUGUGGCCAGUGUGUGUUUUGUUUCUGUUUCUGUGUCCACUGCGGUUUCAGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUC--AGUGGGCACCC--ACAAGUGGUGGAGUAGUGG-AGCUCUA ....(((((((((.......)).)))))))((((((.(((((((.((((.(.....(..((((..(((...((((.......))))...)))..))))...)......).)))))))((((((...--.))))))..((--((.....)))).)))))))-))).... ( -62.70, z-score = -2.46, R) >droSec1.super_1 6157078 133 + 14215200 ----------------UAUGUGAAGCUGCAGCUUCAUCUGCGUGUGGUCAGUGUGUGUUU-----CUGUUUC------------GAUUUCAGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUC--AGUGGGCACCCCCACAAGUAGUGGAGCAGUGGAGGCUCCA ----------------...(((((((....)))))))(((((((.((((.(.........-----((((..(------------.......)..))))..........).)))))))((((((...--.))))))..........))))((((((.......)))))) ( -41.91, z-score = -1.21, R) >droSim1.chr2R 7099880 132 + 19596830 ----------------UAUGUGAAGCUGCAGCUUCAUCUGCGUGUGGUCAGUGUGUGUUU-----CUGUUUC------------GAUUUCAGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUC--AGUGGGCACCCCCACAAGUAGUGGAGUAGUGG-GGCUCCA ----------------...(((((((....)))))))(((((((.((((.(.........-----((((..(------------.......)..))))..........).)))))))((((((...--.))))))..........))))((((((.....-.)))))) ( -40.71, z-score = -1.08, R) >dp4.chr3 8681501 149 - 19779522 -----------CAGCUCCGAGGAGGCUGCAACUCCAUCUGCGUGUGGUCAA-GCAUAGUCC----GAGUUCGAGUCUGAGUCCCAGUCCCAGUCCAUGCCACACAGUCCACGCCUACGUCCAUGUCUAUG-GAGCACCCACAAACAGAGCAGAGCGGAGCGGACUC-- -----------..((((((.((((.......)))).((((((((((((...-((......(----(....)).(.(((.....))).)...))....))))))).......((.....(((((....)))-)))).............))))).))))))......-- ( -44.90, z-score = 0.38, R) >droPer1.super_4 745757 139 - 7162766 ---------UGCAGCCCCGAGGAGGCUGCAACUCCAUCUGCGUGUGGUCAA-GCAUAGUCC----GAGUCCAAGUC------------CCAGUCCAUGCCACACAGUCCACGCCUACGUCCAUGUCAUUG-GAGCACCCACAAACAGAGCAGAGCGGAGCGGACUC-- ---------((((((((....).))))))).((((.((((((((((((...-.....(..(----........)..------------)........))))))).......((.....((((......))-)))).............)))))..)))).......-- ( -42.79, z-score = -0.36, R) >consensus ___________CCGUUUGGAGGAGGCUGCAGCUUCAUCUGCGUGUGGUCAGUGUGUGUUU_____CUGUUUCUGU_U_______GAUUUCAGUUAUAGUUUCUUUUUUCCGGCCCACGCCCAUGUC__AGUGGGCACCCCCACAAGUAGUGGAGCAGUGG_GGCUCCA .....................(((((....))))).((((((((.((((.............................................................)))))))((((((......)))))).............)))))............... (-22.51 = -22.72 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:34 2011