
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,616,982 – 8,617,063 |
| Length | 81 |
| Max. P | 0.976965 |

| Location | 8,616,982 – 8,617,063 |
|---|---|
| Length | 81 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 58.76 |
| Shannon entropy | 0.79313 |
| G+C content | 0.49759 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -5.76 |
| Energy contribution | -5.36 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
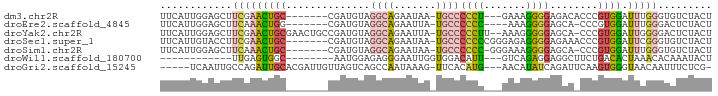
>dm3.chr2R 8616982 81 + 21146708 UUCAUUGGAGCUUCGAACUGC-------CGAUGUAGGCAGAAUAA-UGCCCCCU---GAAAGGGGAGACACCCGUGGAUUUGGGUGUCUACU ..((((((((.......)).)-------)))))..((((......-))))((((---....))))(((((((((......)))))))))... ( -33.90, z-score = -3.73, R) >droEre2.scaffold_4845 5397337 79 - 22589142 UUCAUUGGAGCUUCAAACUGG-------CGAUGUAGGCAGAAUUA-UGCCCCCC----AAAGGGGAGCA-CCCGUGGAUUUGGGACUCUACU .....(((((..(((((.(.(-------((.(((.((((......-))))((((----...)))).)))-..))).).)))))..))))).. ( -25.50, z-score = -0.85, R) >droYak2.chr2R 12805191 88 - 21139217 UUCAUUGGAGCUUCGAACUGCGAACUGCCGAUGUAGGCAGAAUUA-UGCCCCCUU--AAAGGGGGAGCA-CCCGUGGAUUGGGGACUCUACU .....(((((((((((.(..((..(((((......))))).....-((((((((.--...))))).)))-..))..).)))))).))))).. ( -38.30, z-score = -3.96, R) >droSec1.super_1 6155104 84 + 14215200 UUCAUUGUACCUUCGAACUGC-------CGAUGUAGGCAGAAUAA-UGCCCCCCCGGGAGAGGGGAGAAACCCGUGGAUUCGGGUGUCUACU .........(((((...((((-------(......))))).....-..(((....))).))))).(((.(((((......))))).)))... ( -30.70, z-score = -1.62, R) >droSim1.chr2R 7097890 82 + 19596830 UUCAUUGGAGCUUCAAACUGC-------CGAUGUAGGCAGAAUAA-UGCCCCCC-GGGAAAGGGGAGCA-CCCGUGGAUUUGGGUGUCUACU ..((((((((.......)).)-------)))))..((((......-))))((((-......)))).(((-((((......)))))))..... ( -29.80, z-score = -1.39, R) >droWil1.scaffold_180700 6099282 69 - 6630534 ------------UUGAGUGGC--------AAUGGAGAGGGAAUUGGUGGACAUU---GUCAGAGGAGGCUUCUGACACUAAACACAAAUACU ------------...((((.(--------(((.........))))(((.....(---((((((((...))))))))).....)))...)))) ( -16.00, z-score = -1.41, R) >droGri2.scaffold_15245 15199626 82 + 18325388 -----UCAAUUGCCAGAUUGCACGAUUGUUAGUCAGCCAAUAAAG-UUCACAUG---AACAUAUCAGAUUCAAGUGGGUAACAAUUUCUCG- -----....(((((.(((((((....)).)))))...........-..(((.((---(((......).)))).))))))))..........- ( -12.30, z-score = 0.10, R) >consensus UUCAUUGGAGCUUCGAACUGC_______CGAUGUAGGCAGAAUAA_UGCCCCCC___GAAAGGGGAGAA_CCCGUGGAUUUGGGUGUCUACU ................................((((..............((((.......)))).....(((........)))...)))). ( -5.76 = -5.36 + -0.40)
| Location | 8,616,982 – 8,617,063 |
|---|---|
| Length | 81 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 58.76 |
| Shannon entropy | 0.79313 |
| G+C content | 0.49759 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -9.02 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
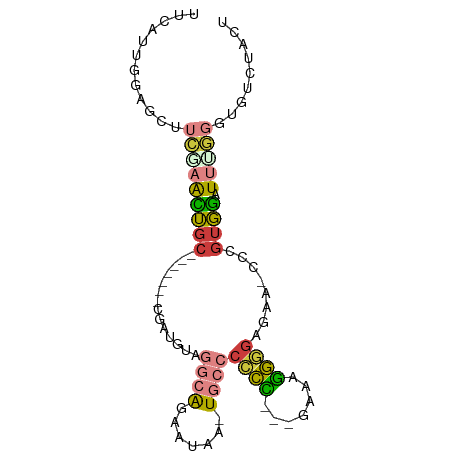
>dm3.chr2R 8616982 81 - 21146708 AGUAGACACCCAAAUCCACGGGUGUCUCCCCUUUC---AGGGGGCA-UUAUUCUGCCUACAUCG-------GCAGUUCGAAGCUCCAAUGAA .(.((((((((........)))))))).)...(((---(.(((((.-((...(((((......)-------))))...)).)))))..)))) ( -31.40, z-score = -3.90, R) >droEre2.scaffold_4845 5397337 79 + 22589142 AGUAGAGUCCCAAAUCCACGGG-UGCUCCCCUUU----GGGGGGCA-UAAUUCUGCCUACAUCG-------CCAGUUUGAAGCUCCAAUGAA .((((((((((........)))-((((((((...----))))))))-..)))))))...(((..-------..(((.....)))...))).. ( -23.40, z-score = -0.54, R) >droYak2.chr2R 12805191 88 + 21139217 AGUAGAGUCCCCAAUCCACGGG-UGCUCCCCCUUU--AAGGGGGCA-UAAUUCUGCCUACAUCGGCAGUUCGCAGUUCGAAGCUCCAAUGAA ....(((..(((.......)))-..)))((((...--..))))((.-.....(((((......)))))((((.....))))))......... ( -27.80, z-score = -1.57, R) >droSec1.super_1 6155104 84 - 14215200 AGUAGACACCCGAAUCCACGGGUUUCUCCCCUCUCCCGGGGGGGCA-UUAUUCUGCCUACAUCG-------GCAGUUCGAAGGUACAAUGAA .(((((.(((((......))))).)))((((((....)))))))).-(((((.(((((...(((-------(....))))))))).))))). ( -29.40, z-score = -1.41, R) >droSim1.chr2R 7097890 82 - 19596830 AGUAGACACCCAAAUCCACGGG-UGCUCCCCUUUCCC-GGGGGGCA-UUAUUCUGCCUACAUCG-------GCAGUUUGAAGCUCCAAUGAA .......((((........)))-)(((((((......-))))))).-(((..(((((......)-------))))..)))............ ( -29.60, z-score = -2.18, R) >droWil1.scaffold_180700 6099282 69 + 6630534 AGUAUUUGUGUUUAGUGUCAGAAGCCUCCUCUGAC---AAUGUCCACCAAUUCCCUCUCCAUU--------GCCACUCAA------------ .......(((..((.(((((((.......))))))---).))..)))................--------.........------------ ( -12.50, z-score = -2.32, R) >droGri2.scaffold_15245 15199626 82 - 18325388 -CGAGAAAUUGUUACCCACUUGAAUCUGAUAUGUU---CAUGUGAA-CUUUAUUGGCUGACUAACAAUCGUGCAAUCUGGCAAUUGA----- -(((......((((.(((..((((..(.((((...---.)))).).-.)))).))).))))......)))(((......))).....----- ( -12.20, z-score = 0.54, R) >consensus AGUAGACACCCAAAUCCACGGG_UGCUCCCCUUUC___AGGGGGCA_UUAUUCUGCCUACAUCG_______GCAGUUCGAAGCUCCAAUGAA .((((...(((........))).....((((((.....))))))............))))................................ ( -9.02 = -8.47 + -0.54)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:33 2011