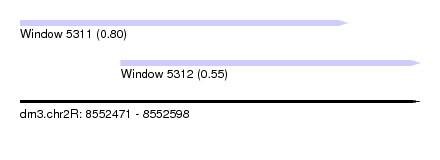
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,552,471 – 8,552,598 |
| Length | 127 |
| Max. P | 0.795900 |

| Location | 8,552,471 – 8,552,575 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.55216 |
| G+C content | 0.45528 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.29 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.795900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
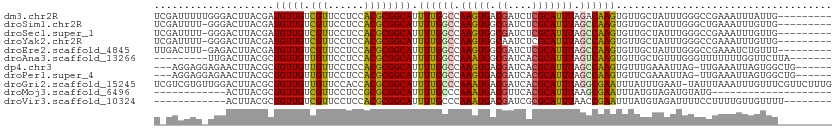
>dm3.chr2R 8552471 104 + 21146708 UCGAUUUUUGGGACUUACGAUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGACGAUCUCGCAUUUAGACAAGUGUUGCUAUUUGGGCCGAAAUUUAUUG--------- (((......((((...(((((...))))))))).....))).((((((((((((((.((....)))))))...((((((....)))))))))))))))......--------- ( -26.10, z-score = -0.37, R) >droSim1.chr2R 7038470 103 + 19596830 UCGAUUUU-GGGACUUACGAUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGUUGCUAUUUGGGCUGAAAUUUGUUG--------- ..(((((.-.(.....(((((...)))))(((.....(((((((((.(((.(((((.((....))))))).)))))))))))).....))))..))))).....--------- ( -29.90, z-score = -0.88, R) >droSec1.super_1 6098835 103 + 14215200 UCGAUUUU-GGGACUUACGAUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGUUGCUAUUUGGGCCGAAAUUUGUUG--------- ..((((((-((.....(((((...)))))(((.....(((((((((.(((.(((((.((....))))))).)))))))))))).....))))))))))).....--------- ( -32.40, z-score = -1.47, R) >droYak2.chr2R 12747349 103 - 21139217 UCGAUUUU-GGGACUUACGAUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGGCAAUCUCGCAUUUAGCCAAGUGUUGCUAUUUGGGCCGAAAUUUGUUG--------- ........-.(((...(((((...)))))...)))..((((.(((((((((((((((((((((.((.....))..)).)))))))))).)))))))))))))..--------- ( -32.50, z-score = -1.73, R) >droEre2.scaffold_4845 5341356 103 - 22589142 UUGACUUU-GAGACUUACGAUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGUUGCUAUUUGGGCCGAAAUCUGUUU--------- ........-(((....(((((...)))))..)))...((((.(((((((((((((((((((((.((.....))..)).)))))))))).)))))))))))))..--------- ( -32.50, z-score = -1.84, R) >droAna3.scaffold_13266 13144058 97 + 19884421 ---------UUGACUUACGCUGUUGUUGUUCCUCCACGCGGCAUUUUGGCCAAAUGGCGAUCACGCAUUUAGUCAAGUGUUGCUGUUUGGGUUUUUUUGGUUCUUA------- ---------.......................((((.((((((.((((((.(((((.((....))))))).))))))...)))))).))))...............------- ( -23.20, z-score = -0.10, R) >dp4.chr3 8624493 103 - 19779522 ---AGGAGGAGAACUUACGCUGUUGUUGUUCCUCCACGCGGCAUUUUGGCCAAGUGACGAUCACGCAUUUAGCCAAGUGUUUGAAAUUAG-UUGAAAUUAGUGGCUG------ ---.((((((((((..........))).)))))))..(((((......)))..(((.....)))))...((((((...((((.((.....-)).))))...))))))------ ( -31.50, z-score = -1.75, R) >droPer1.super_4 686656 103 - 7162766 ---AGGAGGAGAACUUACGCUGUUGUUGUUCCUCCACGCGGCAUUUUGGCCAAGUGACGAUCACGCAUUUAGCCAAGUGUUCGAAAUUAG-UUGAAAUUAGUGGCUG------ ---.((((((((((..........))).)))))))..(((((......)))..(((.....)))))...((((((....(((((......-))))).....))))))------ ( -32.70, z-score = -2.04, R) >droGri2.scaffold_15245 15117058 112 + 18325388 UCGUCGUGUUGGACUUACGCUGUUGUUGUUCCACCACGCGGCAUUUUGCCCAAAUGACGAUCACGCAUUUAGGCGAAUUUAUUUGAAU-UAUUUAAAUUUGUUUCGUUCUUUG ..(((((((((((...(((....)))...))))..))))))).....(((.(((((.((....))))))).)))(((...((((((..-...))))))....)))........ ( -26.30, z-score = -1.08, R) >droMoj3.scaffold_6496 24540006 81 - 26866924 ------------ACUUACGCUGUUGUCGUUCCUCCGCGCGGCAUUUUGCCCAAAUGACGUUCACGCAUUUAAGCGAAUUUAUGUAGAUGUAUG-------------------- ------------...(((((((.((((((........)))))).(((((..(((((.((....)))))))..)))))......))).))))..-------------------- ( -17.20, z-score = 0.04, R) >droVir3.scaffold_10324 788925 93 - 1288806 ------------ACUUACGCUGUUGUCGUUCCUCCACGCGGCAUUUUGCCCAAAUGACGAUCGCGCAUUUAACCGAAUUUAUGUAGAUUUUCCUUUUGUUGUUUU-------- ------------.....(((.((((((((......))(.(((.....))))....)))))).)))........................................-------- ( -14.70, z-score = 0.32, R) >consensus U_GAUUUU_GGGACUUACGCUGUUGUCGUUCCUCCACGCGGCAUUUUGGCCAAGUGACGAUCACGCAUUUAGCCAAGUGUUGCUAUUUGGGCCGAAAUUUGUUG_________ ....................(((((.(((......)))))))).((((((.(((((.((....))))))).)))))).................................... (-16.06 = -15.29 + -0.77)

| Location | 8,552,503 – 8,552,598 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.58867 |
| G+C content | 0.45105 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -13.10 |
| Energy contribution | -11.84 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8552503 95 + 21146708 UCCACGCGGCAUUUUGGCCAAGUGACGAUCUCGCAUUUAGACAAGUGU-UGCUAUUUGGGCCGAAAUUUAUUGGGUUUUUC---UUGCUUAUGCUGUCA------ .....(((((((((((((((((((.((....)))))))...((((((.-...)))))))))))))).....(((((.....---..)))))))))))..------ ( -27.00, z-score = -1.29, R) >droSim1.chr2R 7038501 95 + 19596830 UCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGU-UGCUAUUUGGGCUGAAAUUUGUUGGGUUUUUC---UUGCUCACACUGUCA------ .....(((((((((.(((.(((((.((....))))))).)))))))))-)))....(((((.((((...........))))---..)))))........------ ( -28.50, z-score = -1.05, R) >droSec1.super_1 6098866 95 + 14215200 UCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGU-UGCUAUUUGGGCCGAAAUUUGUUGGGUUUUUC---UUGCUUACGCUGUCA------ .....((((((((((((((((((((((((((.((.....))..)).))-)))))))).)))))))))....(((((.....---..))))).)))))..------ ( -31.30, z-score = -1.59, R) >droYak2.chr2R 12747380 97 - 21139217 UCCACGCGGCAUUUUGGCCAAGUGGCAAUCUCGCAUUUAGCCAAGUGU-UGCUAUUUGGGCCGAAAUUUGUUGGGUUUUUCGCUUCGCUCGC-CUGUGA------ ..((((.((((((((((((((((((((((((.((.....))..)).))-)))))))).)))))))))..((..(((.....)))..))..))-))))).------ ( -35.00, z-score = -2.00, R) >droEre2.scaffold_4845 5341387 94 - 22589142 UCCACGCGGCAUUUUGGCCAAGUGGCGAUCUCGCAUUUAGCCAAGUGU-UGCUAUUUGGGCCGAAAUCUGUUUGGUUUUUC---C-GCUUACGCUGUCA------ .(((.((((.(((((((((((((((((((((.((.....))..)).))-)))))))).))))))))))))).)))......---.-((....)).....------ ( -33.20, z-score = -2.31, R) >dp4.chr3 8624522 97 - 19779522 UCCACGCGGCAUUUUGGCCAAGUGACGAUCACGCAUUUAGCCAAGU--GUUUGAAAUUAGUUGAAAUUAGUGGCUGGGUUUCGCUUCUUGUUUUUGUCA------ .....(((((......)))..(((.....)))))(((((((((...--((((.((.....)).))))...)))))))))....................------ ( -22.00, z-score = 0.47, R) >droPer1.super_4 686685 97 - 7162766 UCCACGCGGCAUUUUGGCCAAGUGACGAUCACGCAUUUAGCCAAGU--GUUCGAAAUUAGUUGAAAUUAGUGGCUGGGUUUCGCUUCUUGUUUUUGUCA------ .....(((((......)))..(((.....)))))(((((((((...--.(((((......))))).....)))))))))....................------ ( -23.20, z-score = 0.12, R) >droGri2.scaffold_15245 15117090 103 + 18325388 ACCACGCGGCAUUUUGCCCAAAUGACGAUCACGCAUUUAGGCGAAUUUAUUUGAAU-UAUUUAAAUU-UGUUUCGUUCUUUGGUUUUUGGUUUUUGUUUUUACUU ((((...(((.....)))((((.(((((...(((......)))(((..((((((..-...)))))).-.)))))))).)))).....)))).............. ( -19.10, z-score = -0.88, R) >droMoj3.scaffold_6496 24540026 96 - 26866924 UCCGCGCGGCAUUUUGCCCAAAUGACGUUCACGCAUUUAAGCGAAUUUAUGUAGAUGUAUGUGCCUUGUGUUUAGCUUUUGUUUCUUUCGCGCUCG--------- ...((((((((((((((..(((((.((....)))))))..)))))..(((......))).)))))..(.....(((....))).....)))))...--------- ( -21.40, z-score = 0.19, R) >consensus UCCACGCGGCAUUUUGGCCAAGUGACGAUCUCGCAUUUAGCCAAGUGU_UGCUAUUUGGGCUGAAAUUUGUUGGGUUUUUC___UUGCUGACGCUGUCA______ ......((((..((((((.(((((.((....))))))).))))))......((....)))))).......................................... (-13.10 = -11.84 + -1.25)

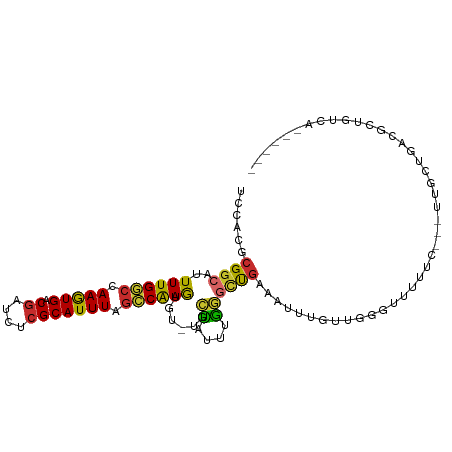
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:28 2011