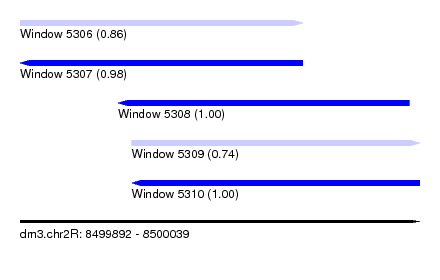
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,499,892 – 8,500,039 |
| Length | 147 |
| Max. P | 0.997952 |

| Location | 8,499,892 – 8,499,996 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.35677 |
| G+C content | 0.40618 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8499892 104 + 21146708 CCUUAUUUUUAAGGAUGUCUGGCGUUGCCAUCCUUAUUUG-AAGGAUGUAAGGCGUUGCCAACCUUAUUGCUAAGAAUGUGUAUUGACACACAUGAAUAGAAAUC (((((....)))))..(..(((((.((((((((((.....-))))))....)))).)))))..)......(((...((((((......))))))...)))..... ( -29.00, z-score = -2.78, R) >droSim1.chr2R 6984470 104 + 19596830 -CCUUUUUCUAAGGAUAUACGGCGUUGCCAUCCUUAUUUUUAAGGACGUCAGGCGUUGCCGUCGUAAAUGCCAAGAAUGUGUUUUGACACACAUGUGUAGAGUUC -.....(((((.((.(((((((((.((((.((((((....)))))).....)))).)))))).)))....))....((((((......))))))...)))))... ( -29.60, z-score = -2.23, R) >droSec1.super_1 6045996 105 + 14215200 CCUUUUUUCUAAGGAUAUACGGCGUUGCCAUCCUUAUUUUUAAGGACGUCAGGCGUUGCCGUCCUAAAUGCUAAGAAUGUGUAUUGACACACAUGUGUAGAGUUC ((((......))))....((((((.((((.((((((....)))))).....)))).))))))........(((...((((((......))))))...)))..... ( -30.60, z-score = -2.78, R) >droYak2.chr2R 12694936 103 - 21139217 CCUGAUUUCUAAGGAUUUCGGACGCUACCACUUUAUUUGUUAAGAAUGCCGGGUGUUGCCAUCACAAUUCCUAAG--UUUGUAUUGAUACACAUGUGUAGAAAUC ...((((((((((((.....((.((...((((((((((.....)))))..)))))..))..)).....))))...--..((((....))))......)))))))) ( -18.20, z-score = 0.58, R) >consensus CCUUAUUUCUAAGGAUAUACGGCGUUGCCAUCCUUAUUUUUAAGGACGUCAGGCGUUGCCAUCCUAAAUGCUAAGAAUGUGUAUUGACACACAUGUGUAGAAAUC ......(((((........(((((.(((((((((((....)))))))....)))).)))))...............((((((......))))))...)))))... (-18.30 = -19.18 + 0.88)

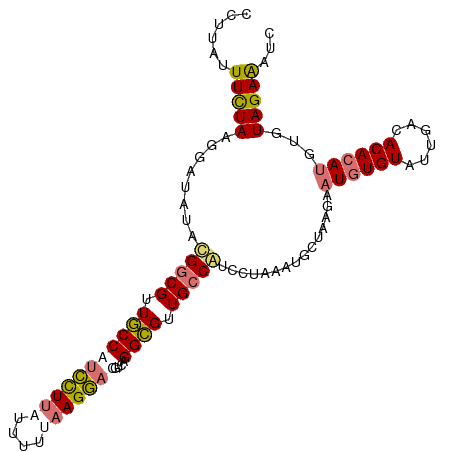
| Location | 8,499,892 – 8,499,996 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.35677 |
| G+C content | 0.40618 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8499892 104 - 21146708 GAUUUCUAUUCAUGUGUGUCAAUACACAUUCUUAGCAAUAAGGUUGGCAACGCCUUACAUCCUU-CAAAUAAGGAUGGCAACGCCAGACAUCCUUAAAAAUAAGG (((.(((....(((((((....))))))).....((..((((((.(....)))))))(((((((-.....)))))))))......))).)))((((....)))). ( -26.70, z-score = -3.39, R) >droSim1.chr2R 6984470 104 - 19596830 GAACUCUACACAUGUGUGUCAAAACACAUUCUUGGCAUUUACGACGGCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAAAAGG- ............(((((......)))))((((.((.((.....(((((...(((....(((((((....))))))))))...)))))..))))..)))).....- ( -26.70, z-score = -2.48, R) >droSec1.super_1 6045996 105 - 14215200 GAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAUUUAGGACGGCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAAAAAGG ......(((..(((((((....))))))).....((..(((((.((....)))))))((((((((....))))))))))......)))...((((......)))) ( -28.80, z-score = -3.32, R) >droYak2.chr2R 12694936 103 + 21139217 GAUUUCUACACAUGUGUAUCAAUACAAA--CUUAGGAAUUGUGAUGGCAACACCCGGCAUUCUUAACAAAUAAAGUGGUAGCGUCCGAAAUCCUUAGAAAUCAGG ((((((((....(((((....)))))..--...((((.......((....))..(((.((.((..((.......))...)).)))))...))))))))))))... ( -19.90, z-score = -0.57, R) >consensus GAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAAUUAGGACGGCAACGCCUGACAUCCUUAAAAAUAAGGAUGGCAACGCCGGAUAUCCUUAGAAAAAAGG ((((.(((...(((((((....)))))))...))).))))...(((((...(((....((((((......)))))))))...))))).................. (-17.66 = -17.47 + -0.19)


| Location | 8,499,928 – 8,500,035 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.35612 |
| G+C content | 0.45496 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.92 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.997952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8499928 107 - 21146708 UGCCACCCCCACCACAACACAUGUCG-GUGUUGCCAUACGGAUUUCUAUUCAUGUGUGUCAAUACACAUUCUUAGCAAUAAGGUUGGCAACGCCUUACAUCCUUCAAA ....................((((.(-(((((((((..(..(((.(((...(((((((....)))))))...))).)))..)..))))))))))..))))........ ( -29.70, z-score = -3.12, R) >droSim1.chr2R 6984506 103 - 19596830 UGCCAU----ACCUCAACACAUGUCG-GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAAACACAUUCUUGGCAUUUACGACGGCAACGCCUGACGUCCUUAAAA ......----..........((((((-((((((((.....(((..(((...((((((......))))))...)))..))).....))))))))).)))))........ ( -27.70, z-score = -2.39, R) >droSec1.super_1 6046033 107 - 14215200 UGCCAUCCCCACCUCAACACAUGUCG-GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAUUUAGGACGGCAACGCCUGACGUCCUUAAAA ....................((((((-((((((((...(..((..(((...(((((((....)))))))...)))..))..)...))))))))).)))))........ ( -29.60, z-score = -3.11, R) >droYak2.chr2R 12694973 105 + 21139217 UGCCAUCUUAAUUCCAA-GUAGGUCGAGAGCUGCCAUCCCGAUUUCUACACAUGUGUAUCAAUACAAA--CUUAGGAAUUGUGAUGGCAACACCCGGCAUUCUUAACA ((((.((((....((..-...))..))))(.(((((((.(((((((((....(((((....)))))..--..))))))))).))))))).)....))))......... ( -32.40, z-score = -3.90, R) >consensus UGCCAUCCCCACCUCAACACAUGUCG_GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAAUUAGGACGGCAACGCCUGACAUCCUUAAAA ....................((((((.((((((((((...((((.(((...(((((((....)))))))...))).))))...)))))))))).))))))........ (-21.99 = -23.92 + 1.94)


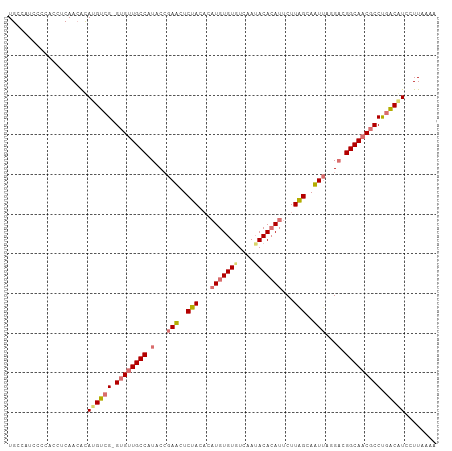
| Location | 8,499,933 – 8,500,039 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Shannon entropy | 0.36121 |
| G+C content | 0.46652 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -24.24 |
| Energy contribution | -26.05 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8499933 106 + 21146708 AGGAUGUAAGGCGUUGCCAACCUUAUUGCUAAGAAUGUGUAUUGACACACAUGAAUAGAAAUCCGUAUGGCAACAC-CGACAUGUGUUGUGGUGGGGGUGGCAACUA ............((((((..........(((...((((((......))))))...)))..((((.(((.(((((((-......))))))).))).)))))))))).. ( -35.40, z-score = -2.08, R) >droSim1.chr2R 6984511 102 + 19596830 AGGACGUCAGGCGUUGCCGUCGUAAAUGCCAAGAAUGUGUUUUGACACACAUGUGUAGAGUUCGGUAUGGCAACAC-CGACAUGUGUUGAGGU----AUGGCAUCUA .(((.((((.((((((((((((....(((.....((((((......))))))..))).....))..))))))))..-((((....))))..))----.)))).))). ( -29.00, z-score = 0.14, R) >droSec1.super_1 6046038 106 + 14215200 AGGACGUCAGGCGUUGCCGUCCUAAAUGCUAAGAAUGUGUAUUGACACACAUGUGUAGAGUUCGGUAUGGCAACAC-CGACAUGUGUUGAGGUGGGGAUGGCAUCUA .(((.((((((.((((((((.(..(((.(((...((((((......))))))...))).)))..).)))))))).)-)..(((.(....).)))....)))).))). ( -35.10, z-score = -1.22, R) >droYak2.chr2R 12694978 104 - 21139217 AGAAUGCCGGGUGUUGCCAUCACAAUUCCUAAG--UUUGUAUUGAUACACAUGUGUAGAAAUCGGGAUGGCAGCUCUCGACCUAC-UUGGAAUUAAGAUGGCAACUC ............(((((((((..((((((.(((--(..(((((..((((....))))..)))(((((.......)))))))..))-))))))))..))))))))).. ( -32.60, z-score = -2.40, R) >consensus AGGACGUCAGGCGUUGCCAUCCUAAAUGCUAAGAAUGUGUAUUGACACACAUGUGUAGAAAUCGGUAUGGCAACAC_CGACAUGUGUUGAGGUGGGGAUGGCAACUA .((((((((...((((((((.((.(((.(((...((((((......))))))...))).))).)).))))))))...((((....)))).........)))))))). (-24.24 = -26.05 + 1.81)

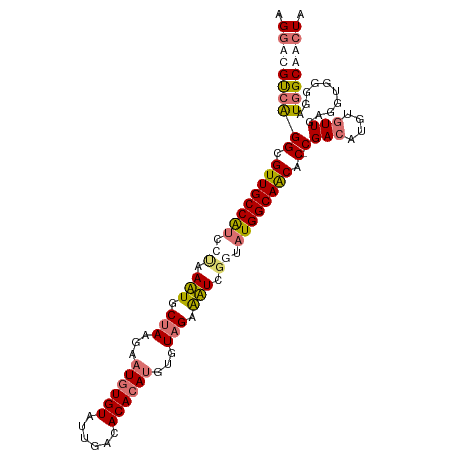
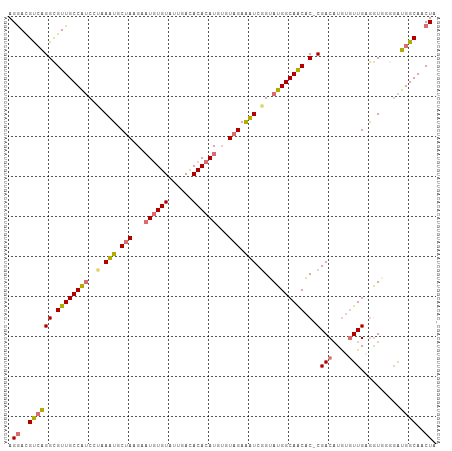
| Location | 8,499,933 – 8,500,039 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Shannon entropy | 0.36121 |
| G+C content | 0.46652 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.92 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8499933 106 - 21146708 UAGUUGCCACCCCCACCACAACACAUGUCG-GUGUUGCCAUACGGAUUUCUAUUCAUGUGUGUCAAUACACAUUCUUAGCAAUAAGGUUGGCAACGCCUUACAUCCU ..((((............))))..((((.(-(((((((((..(..(((.(((...(((((((....)))))))...))).)))..)..))))))))))..))))... ( -31.20, z-score = -3.25, R) >droSim1.chr2R 6984511 102 - 19596830 UAGAUGCCAU----ACCUCAACACAUGUCG-GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAAACACAUUCUUGGCAUUUACGACGGCAACGCCUGACGUCCU (((((((((.----.............(((-((((....))))))).........((((((......))))))...))))))))).((((.((.....)).)))).. ( -28.90, z-score = -2.30, R) >droSec1.super_1 6046038 106 - 14215200 UAGAUGCCAUCCCCACCUCAACACAUGUCG-GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAUUUAGGACGGCAACGCCUGACGUCCU ........................((((((-((((((((...(..((..(((...(((((((....)))))))...)))..))..)...))))))))).)))))... ( -29.60, z-score = -2.51, R) >droYak2.chr2R 12694978 104 + 21139217 GAGUUGCCAUCUUAAUUCCAA-GUAGGUCGAGAGCUGCCAUCCCGAUUUCUACACAUGUGUAUCAAUACAAA--CUUAGGAAUUGUGAUGGCAACACCCGGCAUUCU ..(((((((((.(((((((..-((((((((.((.......)).))))..))))...(((((....)))))..--....))))))).)))))))))............ ( -35.10, z-score = -4.27, R) >consensus UAGAUGCCAUCCCCACCUCAACACAUGUCG_GUGUUGCCAUACCGAACUCUACACAUGUGUGUCAAUACACAUUCUUAGCAAUUAGGACGGCAACGCCUGACAUCCU ........................((((((.((((((((((...((((.(((...(((((((....)))))))...))).))))...)))))))))).))))))... (-21.99 = -23.92 + 1.94)


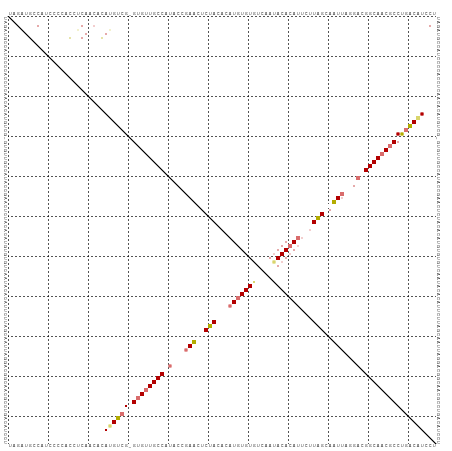
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:26 2011