
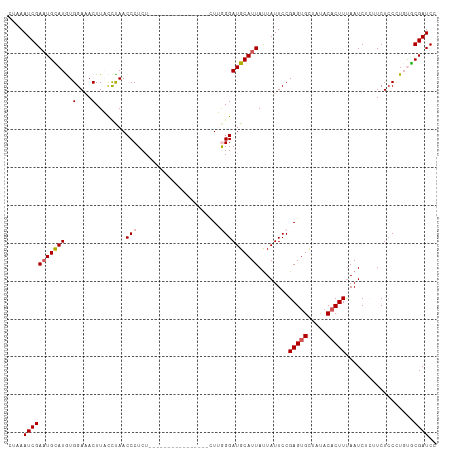
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,438,815 – 8,439,020 |
| Length | 205 |
| Max. P | 0.748417 |

| Location | 8,438,815 – 8,438,912 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Shannon entropy | 0.33647 |
| G+C content | 0.41215 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8438815 97 + 21146708 CUAAAUCGAAUGCAUGUGGAAACUUACCUAACCCUCU----------------UUUGGGAUGCAUUAUUAUUCCGAGUGUUAUACACUUUAAUCUCUUCUCCCUGUGCGAUCC ....(((((((((((..(....)........(((...----------------...))))))))))........(((((.....)))))..................)))).. ( -19.50, z-score = -2.26, R) >droEre2.scaffold_4845 5229360 111 - 22589142 CUAAAUCGAAUGUAUGUGGAAACUUCCCCAACCCUCUGGUGACUUUUAAAAGCCUGAGGAUGCAUUAUUAUUCCGAGUGUUACACACUUUAAUCU--UCUGCUUCCGCGAUCC ....(((((((((((.(((........)))..((((.(((...........))).)))))))))))........(((((.....)))))......--..........)))).. ( -22.30, z-score = -1.41, R) >droYak2.chr2R 12634185 111 - 21139217 CUUAAUCGAUUGUAUGUGGAAACUUCCCUAACCCUCUGAAGGCUUUUAAAAGCCUGAGGAUGCAUUACUAUUCCGAGCGCUACACACUUCAAUCU--UCUCCUUCCACGAUCC .......(((((..((((..............((((...((((((....))))))))))..((.............))....))))...))))).--................ ( -21.12, z-score = -1.34, R) >droSec1.super_1 5985458 97 + 14215200 UUAAAUCGAAUGCAUAUGGAUACUUAUUUAACCCUCU----------------CUUGGGAUGCAUUAUUAUUCCGAGUGCUAUACACUUUAAUCUCUUCUCCCUGUGCGAUCC .......((.((((((.(((...........(((...----------------...)))...............(((((.....)))))..........))).)))))).)). ( -19.20, z-score = -2.23, R) >droSim1.chr2R 6922070 97 + 19596830 CUAAAUCGAAUGCAUAUGGAUACUUACCUAACCCUCU----------------CUUGGGAUGCAUUAUUAUUCCGAGUGCUAUACACUUUAAUCUCUUCUCCCUGUGCGAUCC .......((.((((((.(((...........(((...----------------...)))...............(((((.....)))))..........))).)))))).)). ( -19.20, z-score = -2.39, R) >consensus CUAAAUCGAAUGCAUGUGGAAACUUACCUAACCCUCU________________CUUGGGAUGCAUUAUUAUUCCGAGUGCUAUACACUUUAAUCUCUUCUCCCUGUGCGAUCC .......((.((((((.(((....................................((((((......))))))(((((.....)))))..........))).)))))).)). (-11.90 = -12.86 + 0.96)


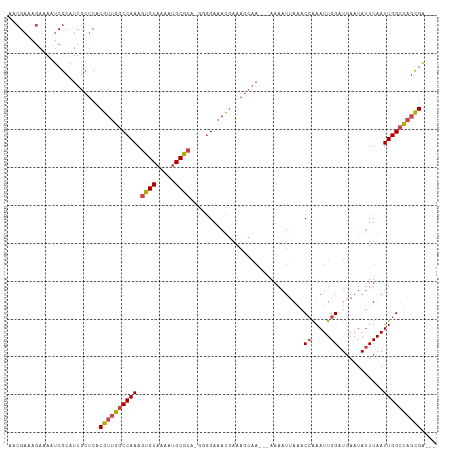
| Location | 8,438,912 – 8,439,020 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Shannon entropy | 0.29040 |
| G+C content | 0.37775 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8438912 108 - 21146708 AACGAAAGAAAAUCGCAUUGCUCACGUUGGCCAAAGUGCAAAAUGCGCA-GGGGAAAUGAAAGCAA-AAAAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCCAGCGA--- ..(....)....((((.(((((..((((..((...((((.....)))).-.))..))))..)))))-...........((((.((((((....))))))))))...))))--- ( -26.40, z-score = -1.79, R) >droAna3.scaffold_13266 13037021 100 - 19884421 --GGUGAAAUAUUCGCAUUUGCCACGUUGGCCAAAGUGCAAAUGGCA---ACAGACAUAAA--------AAAUGGAUGCGGAAUUGGAUGAAUACUUAAUUGGCUUCUGGCUA --.((((.....))))....((((.(..((((((..(((.....)))---.((..(((...--------..)))..)).....................)))))).))))).. ( -20.80, z-score = 0.41, R) >droEre2.scaffold_4845 5229471 110 + 22589142 AACGAAAGAAAAUCGCAUUGCUCACGUUGGCCAAAGUGCAAAAUGCGCAGGAGGAAAUGAAAGCAAAAAAAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCUAGCGA--- ..(....)....((((.(((((..((((..((...((((.....))))....)).))))..)))))............((((.((((((....))))))))))...))))--- ( -24.40, z-score = -1.59, R) >droYak2.chr2R 12634296 106 + 21139217 AACGAAAGAAAAUCGCAUUGCUCACGUUGGCCAAAGUGCAAAAUGCGCA-GAGGAAACGAAAGCAA---AAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCUAGCGA--- ..(....)....((((.(((((..((((..((...((((.....)))).-..)).))))..)))))---.........((((.((((((....))))))))))...))))--- ( -26.90, z-score = -2.53, R) >droSec1.super_1 5985555 106 - 14215200 AACGAAAGAAAAUCGCAUUGCUCACGUUGACCAAAGUGCAAAAUGCGCA-GGGGAAAUGAAAGCAA---AAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCUAGUGA--- ..(....)....((((.(((((..((((..((...((((.....)))).-.))..))))..)))))---.........((((.((((((....))))))))))...))))--- ( -26.10, z-score = -2.72, R) >droSim1.chr2R 6922167 106 - 19596830 AACGAAAGAAAAUCGCAUUGCUCACGUUGGCCAAAGUGCAAAAUGCGCA-GGGGAAAUGAAAGCAA---AAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCUAGCGA--- ..(....)....((((.(((((..((((..((...((((.....)))).-.))..))))..)))))---.........((((.((((((....))))))))))...))))--- ( -26.40, z-score = -2.08, R) >consensus AACGAAAGAAAAUCGCAUUGCUCACGUUGGCCAAAGUGCAAAAUGCGCA_GGGGAAAUGAAAGCAA___AAAAUUAAACCAAAUUGGAUGAAUAUUUAAUUGGCUAGCGA___ ........................((((((((((.((((.....))))..............................((.....))............)))))))))).... (-18.25 = -18.58 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:17 2011