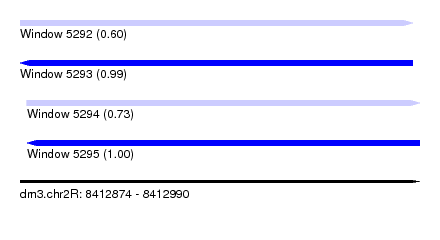
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,412,874 – 8,412,990 |
| Length | 116 |
| Max. P | 0.995243 |

| Location | 8,412,874 – 8,412,988 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.62 |
| Shannon entropy | 0.40438 |
| G+C content | 0.52631 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.70 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8412874 114 + 21146708 CUUUUUAAAUGACUCGCAAUCCGGGUGAGUGGCCCCACAUUGCCCCACCC--AAAGGCCCAAACUUCUGAUCCGCCAUCUG---UGAGUCAGCCUGGCGACCAUGUCAAGUCAUUUCCU ......((((((((.(((....(((((.(.(((........)))))))))--...((((((.....((((..(((.....)---))..))))..))).).)).)))..))))))))... ( -33.80, z-score = -2.27, R) >droAna3.scaffold_13266 13013490 112 + 19884421 -CAUGAAAAUGAGUCGCAAUCUGGGUGAGUGCC-----ACUGCCCCACCCGAAAAAGCCGAAACUUCUGAUCCGCCAUCAGUAGUGAGUCAGCUCUGGGUCUCUUGUGAAUAUUUAUU- -(((....)))..((((((...(((..((....-----.))..)))(((((....(((.((.(((.(((((.....))))).)))...)).))).)))))...)))))).........- ( -29.30, z-score = -1.05, R) >droEre2.scaffold_4845 5203591 101 - 22589142 CUUUGAAA-UGACUCGCAAUCUGGGUGAGUGCCCCCAUAUCGCCCCACCC--AAAGGCCCAAACUUCUG---------------UGAGUCAGUCUUUCGACCCUGUCAAGUCAUUUCCA .(((((..-(((((((((...((((((.(.((.........))).)))))--)(((.......))).))---------------)))))))(((....)))....)))))......... ( -27.00, z-score = -2.42, R) >droYak2.chr2R 12608133 102 - 21139217 CUUCGAAAAUGACUCGCAAUCUGAGUGAGUGCCCCCAAAUCGCCCCACCC--AAAGGCCCAAACUUCUG---------------UGAGUCAGCCUUUCGACCCUGUCAAGUCAUUUCCA ..((((((.(((((((((....((((..(.(((.................--...))).)..)))).))---------------)))))))...))))))................... ( -23.25, z-score = -2.34, R) >droSec1.super_1 5959413 113 + 14215200 CUUUGAAA-UGACUCGCAAUUCGGGUGAGUGGCCGCACAUUGCCCCACCC--AAAGGCCCAAACUUUUGAUCCGCCAUCAG---UGAGUCAGCCUGGCGACCCUGUUAAGUCAUUUCCU ....((((-((((((((.....(((((.(.(((........)))))))))--..((((....(((((((((.....)))))---.))))..)))).)))).........)))))))).. ( -35.20, z-score = -2.04, R) >droSim1.chr2R 6885935 113 + 19596830 CUUUGAAA-UGACUCGCAGUCCGGGUGAGUGGCCCCACAUUGCCCCACCC--AAAGGCCCAAACUUCUGAUCCGCCAUCAG---UGAGUCAGCCGGGCGACCCUGUCAAGUCAUUUCCU ....((((-(((((.((((...(((((.(.(((........)))))))))--....((((......((((..(((.....)---))..))))..))))....))))..))))))))).. ( -41.90, z-score = -3.45, R) >consensus CUUUGAAA_UGACUCGCAAUCCGGGUGAGUGCCCCCACAUUGCCCCACCC__AAAGGCCCAAACUUCUGAUCCGCCAUCAG___UGAGUCAGCCUGGCGACCCUGUCAAGUCAUUUCCU .........((((((.......(((((((((....))).))))))..((......))............................)))))).......(((........)))....... (-13.17 = -14.70 + 1.53)

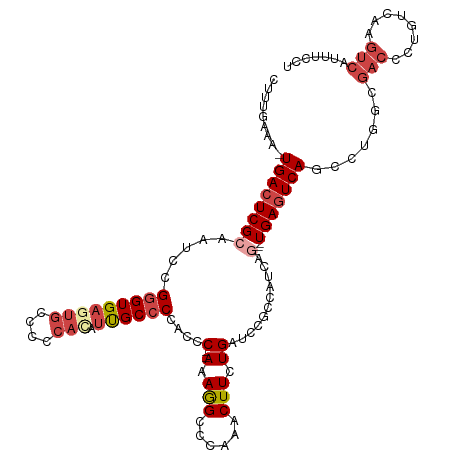
| Location | 8,412,874 – 8,412,988 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Shannon entropy | 0.40438 |
| G+C content | 0.52631 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -23.25 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8412874 114 - 21146708 AGGAAAUGACUUGACAUGGUCGCCAGGCUGACUCA---CAGAUGGCGGAUCAGAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGGGGCCACUCACCCGGAUUGCGAGUCAUUUAAAAAG ...((((((((((.(.(((((((((..(((.....---))).)))).)))))..((((((((((((.--....)))))...(((((....)))))))))))))))))))))))...... ( -43.30, z-score = -2.35, R) >droAna3.scaffold_13266 13013490 112 - 19884421 -AAUAAAUAUUCACAAGAGACCCAGAGCUGACUCACUACUGAUGGCGGAUCAGAAGUUUCGGCUUUUUCGGGUGGGGCAGU-----GGCACUCACCCAGAUUGCGACUCAUUUUCAUG- -...............(((....((((((((...(((.(((((.....))))).))).))))))))...(((((((((...-----.)).))))))).........))).........- ( -33.70, z-score = -1.75, R) >droEre2.scaffold_4845 5203591 101 + 22589142 UGGAAAUGACUUGACAGGGUCGAAAGACUGACUCA---------------CAGAAGUUUGGGCCUUU--GGGUGGGGCGAUAUGGGGGCACUCACCCAGAUUGCGAGUCA-UUUCAAAG ..(((((((((((.((((.(((((...(((.....---------------)))...))))).))(((--(((((((((.........)).)))))))))).)))))))))-)))).... ( -41.80, z-score = -4.42, R) >droYak2.chr2R 12608133 102 + 21139217 UGGAAAUGACUUGACAGGGUCGAAAGGCUGACUCA---------------CAGAAGUUUGGGCCUUU--GGGUGGGGCGAUUUGGGGGCACUCACUCAGAUUGCGAGUCAUUUUCGAAG .(((((((((((.....((((....))))..((((---------------(.((((.......))))--..)))))((((((((((........))))))))))))))))))))).... ( -40.50, z-score = -3.95, R) >droSec1.super_1 5959413 113 - 14215200 AGGAAAUGACUUAACAGGGUCGCCAGGCUGACUCA---CUGAUGGCGGAUCAAAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGCGGCCACUCACCCGAAUUGCGAGUCA-UUUCAAAG ..((((((((((....((..(((((..(.......---..).)))))..))...((((((((....(--((((((.((.....))..)))))))))))))))..))))))-)))).... ( -41.40, z-score = -1.85, R) >droSim1.chr2R 6885935 113 - 19596830 AGGAAAUGACUUGACAGGGUCGCCCGGCUGACUCA---CUGAUGGCGGAUCAGAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGGGGCCACUCACCCGGACUGCGAGUCA-UUUCAAAG ..(((((((((((.(((....(((((...((((..---(((((.....))))).))))))))).(((--(((((((((........))).).))))))))))))))))))-)))).... ( -49.00, z-score = -2.80, R) >consensus AGGAAAUGACUUGACAGGGUCGCAAGGCUGACUCA___CUGAUGGCGGAUCAGAAGUUUGGGCCUUU__GGGUGGGGCAAUGUGGGGCCACUCACCCAGAUUGCGAGUCA_UUUCAAAG .((((.(((((((..((((((...(((((.........................))))).))))))...(((((((..............)))))))......))))))).)))).... (-23.25 = -24.20 + 0.95)

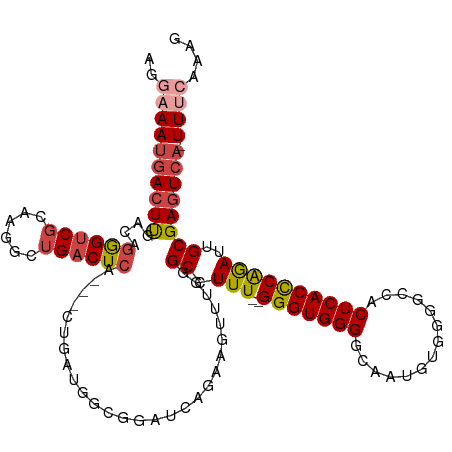
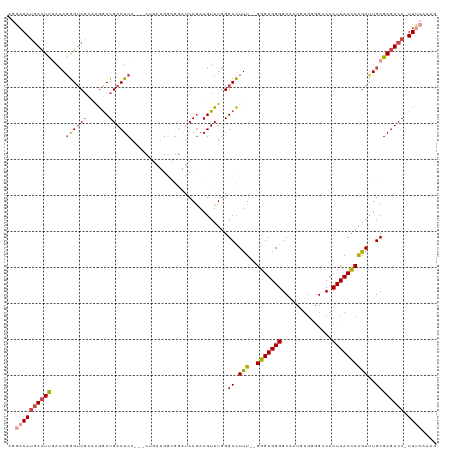
| Location | 8,412,876 – 8,412,990 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Shannon entropy | 0.38631 |
| G+C content | 0.51402 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8412876 114 + 21146708 -UUUUAAAUGACUCGCAAUCCGGGUGAGUGGCCCCACAUUGCCCCACCC--AAAGGCCCAAACUUCUGAUCCGCCAUCUG---UGAGUCAGCCUGGCGACCAUGUCAAGUCAUUUCCUUU -....((((((((.(((....(((((.(.(((........)))))))))--...((((((.....((((..(((.....)---))..))))..))).).)).)))..))))))))..... ( -33.80, z-score = -2.32, R) >droAna3.scaffold_13266 13013491 113 + 19884421 -AUGAAAAUGAGUCGCAAUCUGGGUGAGUGCC-----ACUGCCCCACCCGAAAAAGCCGAAACUUCUGAUCCGCCAUCAGUAGUGAGUCAGCUCUGGGUCUCUUGUGAAU-AUUUAUUUU -...((((((((((((((...(((..((....-----.))..)))(((((....(((.((.(((.(((((.....))))).)))...)).))).)))))...))))))..-.)))))))) ( -29.60, z-score = -1.16, R) >droEre2.scaffold_4845 5203592 102 - 22589142 -UUUGAAAUGACUCGCAAUCUGGGUGAGUGCCCCCAUAUCGCCCCACCC--AAAGGCCCAAACUUCUG---------------UGAGUCAGUCUUUCGACCCUGUCAAGUCAUUUCCAUU -(((((..(((((((((...((((((.(.((.........))).)))))--)(((.......))).))---------------)))))))(((....)))....)))))........... ( -26.90, z-score = -2.48, R) >droYak2.chr2R 12608134 103 - 21139217 UUCGAAAAUGACUCGCAAUCUGAGUGAGUGCCCCCAAAUCGCCCCACCC--AAAGGCCCAAACUUCUG---------------UGAGUCAGCCUUUCGACCCUGUCAAGUCAUUUCCAUU .((((((.(((((((((....((((..(.(((.................--...))).)..)))).))---------------)))))))...))))))..................... ( -23.25, z-score = -2.49, R) >droSec1.super_1 5959414 114 + 14215200 -UUUGAAAUGACUCGCAAUUCGGGUGAGUGGCCGCACAUUGCCCCACCC--AAAGGCCCAAACUUUUGAUCCGCCAUCAG---UGAGUCAGCCUGGCGACCCUGUUAAGUCAUUUCCUUU -...((((((((((((.....(((((.(.(((........)))))))))--..((((....(((((((((.....)))))---.))))..)))).)))).........)))))))).... ( -35.20, z-score = -2.17, R) >droSim1.chr2R 6885936 113 + 19596830 -UUUGAAAUGACUCGCAGUCCGGGUGAGUGGCCCCACAUUGCCCCACCC--AAAGGCCCAAACUUCUGAUCCGCCAUCAG---UGAGUCAGCCGGGCGACCCUGUCAAGUCAUUUCCUU- -...(((((((((.((((...(((((.(.(((........)))))))))--....((((......((((..(((.....)---))..))))..))))....))))..)))))))))...- ( -41.90, z-score = -3.56, R) >consensus _UUUGAAAUGACUCGCAAUCCGGGUGAGUGCCCCCACAUUGCCCCACCC__AAAGGCCCAAACUUCUGAUCCGCCAUCAG___UGAGUCAGCCUGGCGACCCUGUCAAGUCAUUUCCUUU .....((((((((........(((((((((....))).))))))..........(((....((((...................))))..)))..............))))))))..... (-14.03 = -14.64 + 0.61)

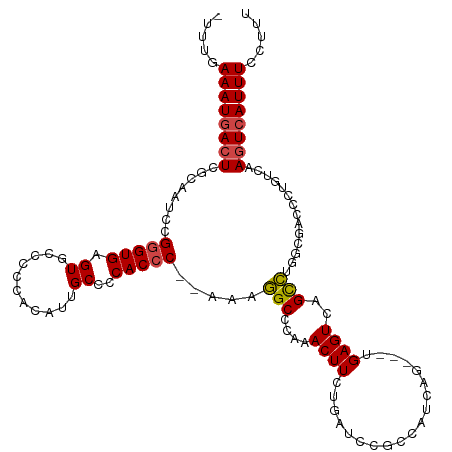
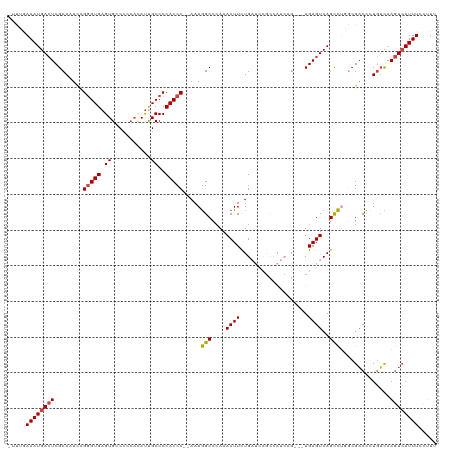
| Location | 8,412,876 – 8,412,990 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Shannon entropy | 0.38631 |
| G+C content | 0.51402 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8412876 114 - 21146708 AAAGGAAAUGACUUGACAUGGUCGCCAGGCUGACUCA---CAGAUGGCGGAUCAGAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGGGGCCACUCACCCGGAUUGCGAGUCAUUUAAAA- .....((((((((((.(.(((((((((..(((.....---))).)))).)))))..((((((((((((.--....)))))...(((((....)))))))))))))))))))))))....- ( -43.30, z-score = -2.33, R) >droAna3.scaffold_13266 13013491 113 - 19884421 AAAAUAAAU-AUUCACAAGAGACCCAGAGCUGACUCACUACUGAUGGCGGAUCAGAAGUUUCGGCUUUUUCGGGUGGGGCAGU-----GGCACUCACCCAGAUUGCGACUCAUUUUCAU- .........-........(((....((((((((...(((.(((((.....))))).))).))))))))...(((((((((...-----.)).))))))).........)))........- ( -33.70, z-score = -1.85, R) >droEre2.scaffold_4845 5203592 102 + 22589142 AAUGGAAAUGACUUGACAGGGUCGAAAGACUGACUCA---------------CAGAAGUUUGGGCCUUU--GGGUGGGGCGAUAUGGGGGCACUCACCCAGAUUGCGAGUCAUUUCAAA- ....(((((((((((.((((.(((((...(((.....---------------)))...))))).))(((--(((((((((.........)).)))))))))).)))))))))))))...- ( -41.80, z-score = -4.28, R) >droYak2.chr2R 12608134 103 + 21139217 AAUGGAAAUGACUUGACAGGGUCGAAAGGCUGACUCA---------------CAGAAGUUUGGGCCUUU--GGGUGGGGCGAUUUGGGGGCACUCACUCAGAUUGCGAGUCAUUUUCGAA ..((((((((((((.....((((....))))..((((---------------(.((((.......))))--..)))))((((((((((........)))))))))))))))))))))).. ( -40.80, z-score = -4.10, R) >droSec1.super_1 5959414 114 - 14215200 AAAGGAAAUGACUUAACAGGGUCGCCAGGCUGACUCA---CUGAUGGCGGAUCAAAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGCGGCCACUCACCCGAAUUGCGAGUCAUUUCAAA- ....((((((((((....((..(((((..(.......---..).)))))..))...((((((((....(--((((((.((.....))..)))))))))))))))..))))))))))...- ( -41.40, z-score = -1.86, R) >droSim1.chr2R 6885936 113 - 19596830 -AAGGAAAUGACUUGACAGGGUCGCCCGGCUGACUCA---CUGAUGGCGGAUCAGAAGUUUGGGCCUUU--GGGUGGGGCAAUGUGGGGCCACUCACCCGGACUGCGAGUCAUUUCAAA- -...(((((((((((.(((....(((((...((((..---(((((.....))))).))))))))).(((--(((((((((........))).).))))))))))))))))))))))...- ( -49.00, z-score = -2.79, R) >consensus AAAGGAAAUGACUUGACAGGGUCGCAAGGCUGACUCA___CUGAUGGCGGAUCAGAAGUUUGGGCCUUU__GGGUGGGGCAAUGUGGGGCCACUCACCCAGAUUGCGAGUCAUUUCAAA_ ....(((((((((((..((((((...(((((.........................))))).))))))...(((((((..............)))))))......))))))))))).... (-26.77 = -27.25 + 0.48)

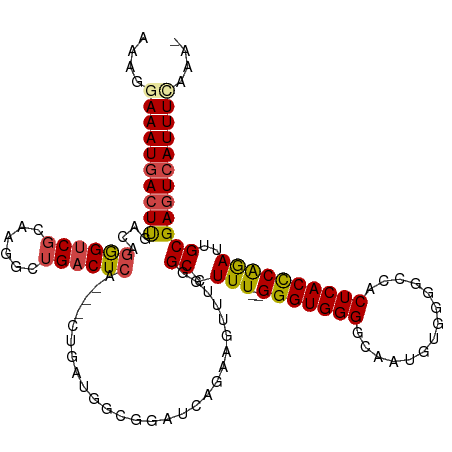
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:14 2011