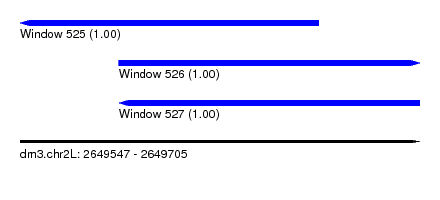
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,649,547 – 2,649,705 |
| Length | 158 |
| Max. P | 0.998978 |

| Location | 2,649,547 – 2,649,665 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.89 |
| Shannon entropy | 0.45938 |
| G+C content | 0.44319 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -19.00 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2649547 118 - 23011544 AAAGUGCUAGUGGUGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCGCCCAAAGCAGCUCCGAAAGCAAUUCUUCAAGAAUUCAAAC- (((((((.((.((.(((((((....((....))...))))))).)))).))))))).........(((((((...........)))))))......(.(((((.....))))))....- ( -26.20, z-score = -2.91, R) >droSim1.chr2L 2609082 118 - 22036055 AAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACUCAAAGCAGCUCCGAAAGCAAUUCUUCAAGAAUUCAAAC- (((((((.((.((((((((((....((....))...)))))))))))).))))))).........(((((((...........)))))))......(.(((((.....))))))....- ( -30.30, z-score = -3.38, R) >droSec1.super_5 817967 118 - 5866729 AAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACUCAAAGCAGCUCCGAAAGCAAUUCUUCGAGAAUUCAAAC- (((((((.((.((((((((((....((....))...)))))))))))).)))))))..........((((((...........))))))((((((........)))).))........- ( -30.80, z-score = -3.03, R) >droYak2.chr2L 2640676 118 - 22324452 AAAGUGCUAGUGGGGAAAAUCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGACCACAAAAGUGGUUCCUCUACCGAAAACAGCUCCGCAAGCAGUUUGUCAAGAAUUCAAGC- (((((((.((.((((((((((....((....))...)))))))))))).)))))))((((((....))))))........((....((((.(....).))))..))............- ( -39.40, z-score = -4.38, R) >droEre2.scaffold_4929 2694603 118 - 26641161 AAAGUGCUAGUGGGGAAAACCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGGCCACAAAAGUUGUUCCUCUACCCAAAACAGCUCCGAAGGCAGUUCUUCAAGAGUUCCAGC- (((((((.((.((((((((((....((....))...)))))))))))).))))))).(((.....(((((((...........))))))).....)))....................- ( -39.60, z-score = -4.01, R) >droAna3.scaffold_12984 57676 98 - 754457 AGAGUGACAGCGAUG----CC--UUUAGAAGUGUGCGAUUCUCCAC---------------AACUAGUUGCUCUUCUAGAUCCAACAGUUCCCAAAAUUGUUCGUUCAUGGCUGAAAAC .......((((.(((----..--((((((((...((((((......---------------....)))))).))))))))...(((((((.....)))))))....))).))))..... ( -23.10, z-score = -1.00, R) >consensus AAAGUGCUAGUGGGGAAAACCAAUUCAAAAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACCCAAAACAGCUCCGAAAGCAAUUCUUCAAGAAUUCAAAC_ (((((((.((.((((((((((....((....))...)))))))))))).))))))).........(((((((...........)))))))........(((((.....)))))...... (-19.00 = -20.92 + 1.92)

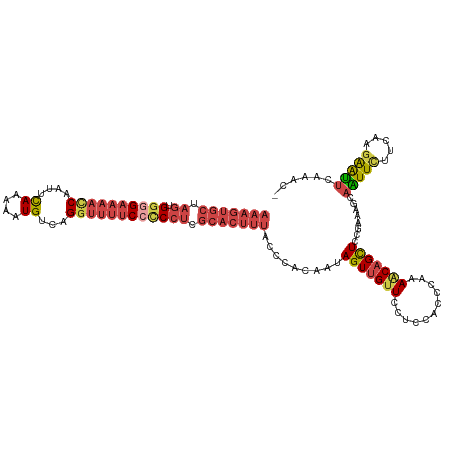
| Location | 2,649,586 – 2,649,705 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Shannon entropy | 0.15714 |
| G+C content | 0.43361 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.62 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2649586 119 + 23011544 GCGGAGGAACAACUAUUGUGGGUAAAGUGCGAGGGGGAAAACCUGACAUUCUUGAAUUGGUUUUCACCACUAGCACUUUCAAAUAUUGUGCUUCCACUACUUUGUCUCGAUACUGAACG ..(((((.((((.((((....(.(((((((.((((.(((((((...((....))....))))))).)).)).)))))))).)))))))).)))))........................ ( -33.10, z-score = -2.04, R) >droSim1.chr2L 2609121 119 + 22036055 GUGGAGGAACAACUAUUGUGGGUAAAGUGCGAGGAGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCGGUUCUAAACG (((((((.((((...((((((...((((((..((.((((((((...((....))....))))))))))....)))))))))))).)))).)))))))...................... ( -39.60, z-score = -3.39, R) >droSec1.super_5 818006 119 + 5866729 GUGGAGGAACAACUAUUGUGGGUAAAGUGCGAGGAGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUAUGCUUCCACUACUUUGUCUCGGUUCUGAACG (((((((........((((((...((((((..((.((((((((...((....))....))))))))))....))))))))))))......)))))))......((.(((....))))). ( -36.44, z-score = -2.54, R) >droYak2.chr2L 2640715 119 + 22324452 GUAGAGGAACCACUUUUGUGGUCAAAGUGCGAGGGGGAAAACCUGACAUUUCUGAAUUGAUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCAAUUCUGAUAG ((((.((((.(((.(((((((...((((((.((((((((((.(...............).)))))))).)).)))))))))))))..))).)))).))))..((((........)))). ( -41.56, z-score = -4.55, R) >droEre2.scaffold_4929 2694642 119 + 26641161 GUAGAGGAACAACUUUUGUGGCCAAAGUGCGAGGGGGAAAACCUGACAUUUCUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCAAUGCAGAAAG ((((.((((((...(((((((...((((((.((((((((((((...............)))))))))).)).)))))))))))))...)).)))).))))(((((......)))))... ( -45.66, z-score = -4.93, R) >consensus GUGGAGGAACAACUAUUGUGGGUAAAGUGCGAGGGGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCGAUUCUGAACG ((((.((((((....((((((...((((((.((((((((((((...((....))....)))))))))).)).))))))))))))....)).)))).))))................... (-33.26 = -34.62 + 1.36)


| Location | 2,649,586 – 2,649,705 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.92 |
| Shannon entropy | 0.15714 |
| G+C content | 0.43361 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -37.02 |
| Energy contribution | -36.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.998978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2649586 119 - 23011544 CGUUCAGUAUCGAGACAAAGUAGUGGAAGCACAAUAUUUGAAAGUGCUAGUGGUGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCGC ......................(((((.(.(((((..((((((((((.((.((.(((((((....((....))...))))))).)))).))))))).....)))..))))).).))))) ( -30.30, z-score = -2.51, R) >droSim1.chr2L 2609121 119 - 22036055 CGUUUAGAACCGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCAC .((((.......))))......(((((.(.(((((((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...)))))..))))).).))))) ( -39.60, z-score = -4.13, R) >droSec1.super_5 818006 119 - 5866729 CGUUCAGAACCGAGACAAAGUAGUGGAAGCAUAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCAC ......................(((((((((....((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...)))))....))))..))))) ( -37.30, z-score = -3.57, R) >droYak2.chr2L 2640715 119 - 22324452 CUAUCAGAAUUGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAAUCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGACCACAAAAGUGGUUCCUCUAC ...................((((.((((.(((..(((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...)))))).))).)))).)))) ( -46.90, z-score = -5.90, R) >droEre2.scaffold_4929 2694642 119 - 26641161 CUUUCUGCAUUGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGGCCACAAAAGUUGUUCCUCUAC .((((......))))....((((.((((.((...(((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...))))))...)))))).)))) ( -45.50, z-score = -4.83, R) >consensus CGUUCAGAAUCGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCAC ......................(((((.(.(((((((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...)))))..))))).).))))) (-37.02 = -36.86 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:58 2011