| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,344,332 – 8,344,433 |
| Length | 101 |
| Max. P | 0.564328 |

| Location | 8,344,332 – 8,344,433 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.60851 |
| G+C content | 0.41750 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
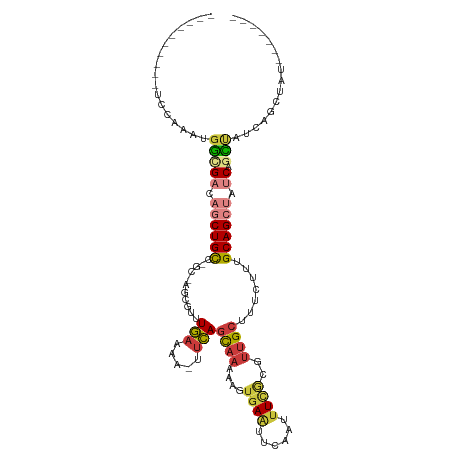
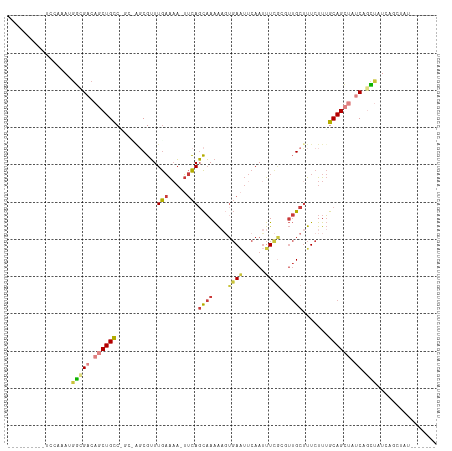
>dm3.chr2R 8344332 101 - 21146708 ------CCAAUACGAAUGGCGACAGCUGCCAGCCAGCGUUUGAAAA-UUCAGCAAAAAGUGAAUUCAAUUUCGCGUUGCUUUCUUUGCAGCUAUCAGCUAUCAGCUAU------- ------.........(((((((.(((((..(((..(((...(((..-...(((((...(((((......))))).))))))))..))).)))..))))).)).)))))------- ( -29.60, z-score = -2.04, R) >dp4.chr3 14036267 89 - 19779522 -----------CCAAUUUUCGACAGCUGUCUGCUGUCGAUUACGAUUGCUAUUUAGGCUUCAG--CAAAUUCA----GCAUUCUUAGCAGAAAACAAACGCUGGCC--------- -----------.......((((((((.....))))))))....((.((((......((....)--)......)----))).))(((((...........)))))..--------- ( -23.20, z-score = -1.58, R) >droAna3.scaffold_13266 454194 84 - 19884421 -----------CCAGAUGUCGGCAGCUGUC----GGCGUUUGAAAAUUUCAGCAAAAUUGGAUUUCAAUUUU--UUUGCUUUCUUUGCAGAAAACAAGCUC-------------- -----------.(((((((((((....)))----))))))))........(((..(((((.....)))))((--(((((.......)))))))....))).-------------- ( -23.80, z-score = -1.89, R) >droEre2.scaffold_4845 5144196 86 + 22589142 ----------CCAAAACGGUGACAGCUGCC----AUCGUUUGAAUA-UUCAGCAAAAAGUGAAUUCAAUUUCGUGUUGCUUUCUUUGCAGCUAUCAGCUAU-------------- ----------.......(.(((.((((((.----......(((...-.)))((((...(((((......))))).)))).......)))))).))).)...-------------- ( -19.30, z-score = -0.96, R) >droYak2.chr2R 12548161 100 + 21139217 ----------UCAAAACGGCGACAGCUGCC----CGCGUUUGAAAA-UUCAGCAAAAAGUGAAUUCAAUUUCGAGUUGCUUUCUUUGCAGCUAUCAGCUAUAAGCUAUCUGGUAU ----------......(((.((.((((((.----.(((..(....)-..).))..(((((.(((((......))))))))))....)))))).))(((.....)))..))).... ( -21.50, z-score = 0.22, R) >droSec1.super_1 5893871 107 - 14215200 CCAAUACGAAUACGAAGGGCGACAGCUGCCAGCCAGCGUUUGAAAA-UUCAGCAAAAAGUGAAUUCAAUUUCGCUUUGCUUUCUUUGCAGCUAUCAGCUAUCAGCUAU------- .................(((((.(((((..(((..(((...(((..-...(((((..((((((......))))))))))))))..))).)))..))))).)).)))..------- ( -27.80, z-score = -1.06, R) >droSim1.chr2R 6821660 107 - 19596830 CCAAUACGAAUACGAAUGGCGACAGCUGCCGGCUAGCGUUUGAAAA-UUCAGCAAAAAGUGAAUUCAAUUUCGCUUUGCUUUCUUUGCAGCUAUCAGCUAUCAGCUAU------- ...............(((((((.(((((..((((.(((...(((..-...(((((..((((((......))))))))))))))..)))))))..))))).)).)))))------- ( -31.60, z-score = -2.19, R) >consensus __________UCCAAAUGGCGACAGCUGCC_GC_AGCGUUUGAAAA_UUCAGCAAAAAGUGAAUUCAAUUUCGCGUUGCUUUCUUUGCAGCUAUCAGCUAUCAGCUAU_______ .................(((((.((((((...........(((.....)))((((....((((......))))..)))).......)))))).)).)))................ ( -9.86 = -10.66 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:18:01 2011