| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,326,012 – 8,326,157 |
| Length | 145 |
| Max. P | 0.929401 |

| Location | 8,326,012 – 8,326,157 |
|---|---|
| Length | 145 |
| Sequences | 6 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 73.53 |
| Shannon entropy | 0.50068 |
| G+C content | 0.39332 |
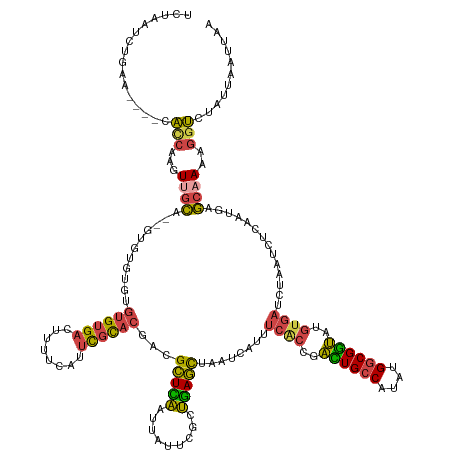
| Mean single sequence MFE | -35.59 |
| Consensus MFE | -21.24 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
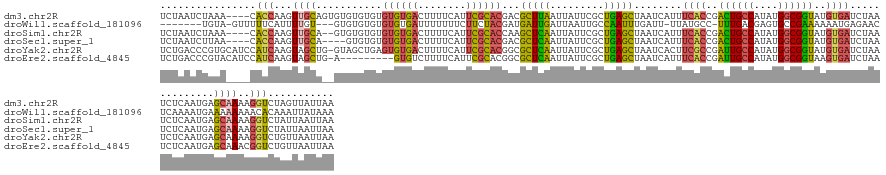
>dm3.chr2R 8326012 145 - 21146708 UCUAAUCUAAA----CACCAAGUUGCAGUGUGUGUGUGUGUGACUUUUCAUUCGCACGACGCUUAAUUAUUCGCUGAGCUAAUCAUUUCACCGACUGCCAUAUGGCGGUAUGUGAUCUAAUCUCAAUGAGCAAAAGGUCUAGUUAUUAA ..(((.(((..----.(((...(((((((((.((((((.((((....)))).))))))))))).......(((.((((.........((((..((((((....))))))..))))......)))).)))))))..))).)))))).... ( -40.06, z-score = -2.18, R) >droWil1.scaffold_181096 1937061 136 + 12416693 -------UGUA-GUUUUUCAUUUUGU---GUGUGUGUGUGUGAUUUUUUUCUUCUACGAUGAUUGAUUAAUUGCCAAUUUGAUU-UUAUGCC-UUUGACGAGUGCCGAAAAAAUGAGAACUCAAAAUGAAAAAAAACACAAAUUAUAAA -------(((.-.((((((((((((.---((.....((.(..(((..((...((......))..))..)))..))).(((.(((-((..(((-((....))).))....))))).))))).))))))))))))..)))........... ( -24.20, z-score = -0.74, R) >droSim1.chr2R 6803403 143 - 19596830 UCUAAUCUAAA----CACCAAGUUGCA--GUGUGUGUGUGUGACUUUUCAUUCGCACCAAGCUCAAUUAUUCGCUGAGCUAAUCAUUUCACCGACUGCCAUAUGGCGGUAUGUGAUCUAAUCUCAAUGAGCAAAAGGUCUAUUAAUUAA ...........----.(((...((((.--.((.(((((.((((....)))).)))))))((((((.........))))))..(((((((((..((((((....))))))..)))).........)))))))))..)))........... ( -37.80, z-score = -2.55, R) >droSec1.super_1 5875382 141 - 14215200 UCUAAUCUUAA----CACCAAGUUGCA----GUGUGUGUGUGACUUUUCAUUCGCACGACGCUCAAUUAUUCGCUGAGCUAAUCAUUUCACCGACUGCCAUAUGGCGGUAUGUGAUCUAAUCUCAAUGAGCAAAAGGUCUAUUAAUUAA ...........----.(((.(((((.(----((((.(((((((........))))))))))))))))).((((.((((.........((((..((((((....))))))..))))......)))).)))).....)))........... ( -37.56, z-score = -2.41, R) >droYak2.chr2R 12529592 148 + 21139217 UCUGACCCGUGCAUCCAUCAAGUAGCUG-GUAGCUGAGUGUGACUUUUCAUUCGCACGGCGCUCAAUUAUUCGCUGAGCUAAUCACUUCGCCGAUUGCCAUAUGGCGGUAUGUGAUCUAAUCUCAAUGAGCAAAAGGUCUGUUAAUUAA ...(((((((((...........(((..-...)))(((((........))))))))))(((((((.........)))))..(((((.((((((.........))))))...))))).............))....)))).......... ( -39.80, z-score = -0.36, R) >droEre2.scaffold_4845 5125611 139 + 22589142 UCUGACCCGUACAUCCAUCAAGUAGCUG-A---------GUGUCUUUUCAUUCGCACGGCGCUCAAUUAUUCGCUGAGCUAAUCAUUUCACCGAUUGCCAUAUGGCGGUAAGUGAUCUAAUCUCAAUGAGCAAACGGUCUGUUAAUUAA ...((((.((...........((.((.(-(---------(((......))))))))).(((((((.........)))))...(((((((((..((((((....))))))..)))).........)))))))..)))))).......... ( -34.10, z-score = -1.04, R) >consensus UCUAAUCUGAA____CACCAAGUUGCA__GUGUGUGUGUGUGACUUUUCAUUCGCACGACGCUCAAUUAUUCGCUGAGCUAAUCAUUUCACCGACUGCCAUAUGGCGGUAUGUGAUCUAAUCUCAAUGAGCAAAAGGUCUAUUAAUUAA ................(((...((((...........((((((........))))))...(((((.........)))))........((((..((((((....))))))..))))..............))))..)))........... (-21.24 = -20.75 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:56 2011