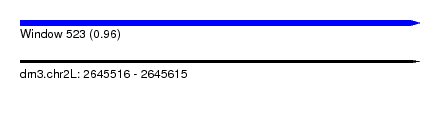
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,645,516 – 2,645,615 |
| Length | 99 |
| Max. P | 0.959555 |

| Location | 2,645,516 – 2,645,615 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 67.40 |
| Shannon entropy | 0.66702 |
| G+C content | 0.53945 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
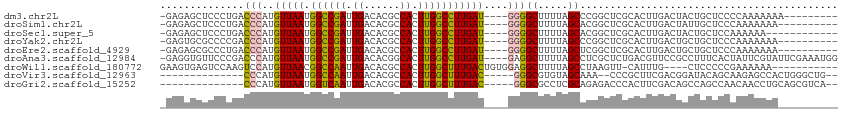
>dm3.chr2L 2645516 99 + 23011544 -GAGAGCUCCCUGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU----GGGGCUUUUAGCCCGGCUCGCACUUGACUACUGCUCCCCAAAAAAA--------- -..(((((...........(((((.((((((.((......)).)))))))))))----.((((.....)))))))))(((.........)))............--------- ( -28.30, z-score = -1.58, R) >droSim1.chr2L 2604957 98 + 22036055 -GAGAGCUCCCUGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU----GGGGCUUUUAGCACGGCUCGCACUUGACUAUUGCUCCCAAAAAAA---------- -..(((((..(((((((.((((((.((((((.((......)).)))))))))))----))))...))))...)))))(((.........)))...........---------- ( -27.10, z-score = -1.21, R) >droSec1.super_5 813787 96 + 5866729 -GAGAGCUCCCUGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU----GGGGCUUUUAGCACGGCUCGCACUUGACUACUGCUCCAAAAAA------------ -..(((((..(((((((.((((((.((((((.((......)).)))))))))))----))))...))))...)))))(((.........))).........------------ ( -27.10, z-score = -1.35, R) >droYak2.chr2L 2635673 98 + 22324452 -GAGUGCGCCCCGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU----GGGGCUUUUAGCCCGGCUCGCACUUGACUGCUGCUCCCAAAAAAA---------- -(((((((..((.......(((((.((((((.((......)).)))))))))))----.((((.....))))))..)))))))....................---------- ( -33.70, z-score = -2.28, R) >droEre2.scaffold_4929 2690325 98 + 26641161 -GAGAGCGCCCUGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU----GGGGCUUUUAGCUCGGCUCGCACUUGACUGCUGCUCCCAAAAAAA---------- -(.(((((((....((((..((((.((((((.((......)).)))))))))))----)))((.....))..)))..(((......))).)))).).......---------- ( -31.00, z-score = -1.57, R) >droAna3.scaffold_12984 42395 108 + 754457 -GAGGUGUUCCCGACCCAUGUUAAUGGCCGAUUGACACGGCACUUGGCCUUGAU----GAGGCUUUUAGCCUCGCUCUGACGUUCCGCCUUUCACUAUUCGUAUUCGAAAUGG -((((((....((......(((((.((((((.((......)).)))))))))))----(((((.....))))).......))...))))))...(((((((....)).))))) ( -30.70, z-score = -1.14, R) >droWil1.scaffold_180772 7573182 97 - 8906247 GAAGUGAGUCCAAGUCCAUGUUAACGGCCAAUUGACACGCCACUUGGCUUUGACUGUGGAGGCUUUUAGCCUAAGUU-CAUUUG----CUCCCCCGAAAAAA----------- .(((((((......((((((((((.((((((.((......)).))))))))))).)))))(((.....)))....))-))))).----..............----------- ( -26.30, z-score = -1.36, R) >droVir3.scaffold_12963 8965718 90 - 20206255 --------------CCCAUGUUAAUGGCCAAUUGACACGCCACUUGGCUUUGAC-----GGGCGUGUAGCAAA--CCCGCUUCGACGGAUACAGCAAGAGCCACUGGGCUG-- --------------....(((((((.....))))))).(((...(((((((...-----(((..((...))..--)))(((((....))...))).)))))))...)))..-- ( -26.10, z-score = -0.03, R) >droGri2.scaffold_15252 11825833 92 - 17193109 --------------CCCAUGUUAAUGGUCAAUUGACACGCCACUUGGCUUUGAC-----GGGCGCCUCGCAGAGACCCACUUCGACAGCCAGCCAACAACCUGCAGCGUCA-- --------------....(((((((.....))))))).(((....)))..((((-----((((...(((.((.......)).)))..))).((.........))..)))))-- ( -22.00, z-score = 0.58, R) >consensus _GAGAGCUCCCUGACCCAUGUUAAUGGCCGAUUGACACGCCACUUGGCCUUGAU____GGGGCUUUUAGCACGGCUCGCACUUGACUGCUGCUCCCAAAAAAA__________ ..............(((..(((((.((((((.((......)).)))))))))))....)))((.....))........................................... (-15.44 = -15.02 + -0.42)
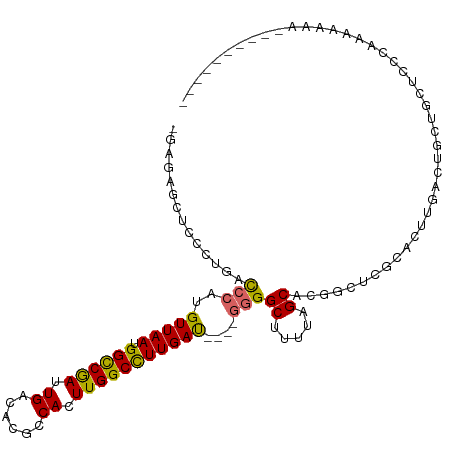

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:55 2011