
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,275,762 – 8,275,865 |
| Length | 103 |
| Max. P | 0.514191 |

| Location | 8,275,762 – 8,275,865 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.08 |
| Shannon entropy | 0.74359 |
| G+C content | 0.45450 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -8.99 |
| Energy contribution | -9.61 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
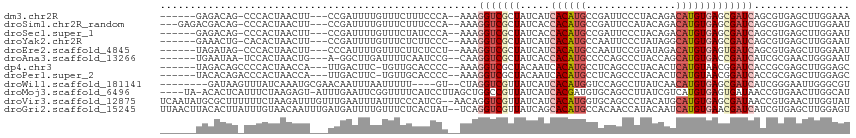
>dm3.chr2R 8275762 103 + 21146708 ------GAGACAG-CCCACUAACUU---CCGAUUUUGUUUCUUUCCCA--AAAGGUCGCUAUCAUCACAUGCCGAUUCCCUACAGACAUGUGAGCGAUCAGCGUGAGCUUGGAAA ------(((((((-...........---......))))))).((((..--...((((((.....(((((((.(...........).)))))))))))))(((....))).)))). ( -27.53, z-score = -2.15, R) >droSim1.chr2R_random 2235824 106 + 2996586 ---GAGACGACAG-CCCACUAACUU---CCGAUUUUGUUUCUUUCCCA--AAAGGUCGCUAUCACCACAUGCCGAUUCCAUACAGACAUGUGAGCGAUCAGCGUGAGCUUGGAAU ---(((((((...-...........---......))))))).((((..--...(((((((.....((((((.(...........).)))))))))))))(((....))).)))). ( -26.85, z-score = -1.90, R) >droSec1.super_1 5820334 103 + 14215200 ------GAGACAG-CCCACUAACUU---CCGAUUUUGUUUCUAUCCCA--AAAGGUCGCUAUCACCACAUGCCGAUUCCCUACAGACAUGUGAGCGAUCAGCGUGAGCUUGGAAU ------(((((((-...........---......)))))))....(((--(..(((((((.....((((((.(...........).))))))))))))).((....))))))... ( -26.83, z-score = -2.18, R) >droYak2.chr2R 12468738 103 - 21139217 ------GAAACUG-CACACUAACUU---CCGAUUUUGUUUCUCUUCCC--AAAGGUCGCUAUCAUCACAUGCCAAUUCCCUAUAGGCAUGUGAGCGAUCAGCGUGAGCUUGGAAU ------(((((..-...........---........)))))..((((.--...((((((.....(((((((((.((.....)).)))))))))))))))(((....))).)))). ( -32.70, z-score = -3.74, R) >droEre2.scaffold_4845 5072125 103 - 22589142 ------UAGAUAG-CCCACUAACUU---CCCAUUUUGUUUCUUCUCCU--AAAGGUCGCUAUCAUCACAUGCCAAUUCCGUAUAGACAUGUGAGUGAUCAGCGUGAGCUUGGAAU ------.......-.........((---((..................--...((((((.....(((((((.(.((.....)).).)))))))))))))(((....))).)))). ( -21.40, z-score = -0.60, R) >droAna3.scaffold_13266 5574063 102 + 19884421 ------UGAAUAA-UCCACUAACUG---A-GGCUUGAUUUUCAAUCCG--CAAGGUCGCUAUCACCACAUGCCCCAGCCCUACCAGCAUGUGACCGAUCAUCGCGAACUGGGAAU ------.......-(((......((---.-((.(((.....))).)).--))((.((((.(((..(((((((.............)))))))...)))....)))).)).))).. ( -21.82, z-score = -0.13, R) >dp4.chr3 7491225 103 + 19779522 ------UAGACAGCCCCACUAACCA---UUGACUUC-UGUUGCACCCC--AAAGGUCGCUACAAUCACAUGCCUCAGCCCUACACUCAUGUAACGGAUCACCGCGAGCUUGGAGC ------..(((((...((.......---.))....)-)))).....((--((.(.((((.......(((((...............)))))...((....)))))).)))))... ( -16.66, z-score = 1.19, R) >droPer1.super_2 7688741 103 + 9036312 ------UACACAGACCCACUAACCA---UUGACUUC-UGUUGCACCCC--AAAGGUCGCUACAAUCACAUGCCUCAGCCCUACACUCAUGUAACGGAUCACCGCGAGCUUGGAGC ------...(((((..((.......---.))...))-)))......((--((.(.((((.......(((((...............)))))...((....)))))).)))))... ( -16.86, z-score = 0.72, R) >droWil1.scaffold_181141 1114899 101 - 5303230 --------GAUAAGUUUAUCAAAUGCGAACAAUUUAAUUUUU----GU--CUAGGUCGUUAUCAUCACAUGGUCCAGCCUUAUCAACAUGUGAGCGAUCAUCGGGAAUUGGGCGU --------((((....))))....(((..((((((.....((----(.--...((((((.....(((((((...............)))))))))))))..)))))))))..))) ( -21.26, z-score = -0.32, R) >droMoj3.scaffold_6496 15062527 109 - 26866924 ----UA-ACACUCAUUUCUAAGAGU-AUUUGAAUUCGGUUUUCAUCCUUAGCUGGCCGUUAUCAUCACGAUGUGCAGCCUUAUCGUCAUGUGAGUGAUAACCGUGAACUUGGCAU ----..-..((((........))))-.........(((((.........)))))((((((((((((((((((...........))))..)))).))))))).(....)..))).. ( -22.70, z-score = 0.12, R) >droVir3.scaffold_12875 2431374 113 - 20611582 UCAAUAUGCGCUUUUUUCUAAGAUUUGUUUGAAUUUAUUUCCCAUCG--AACAGGUCGUUAUCAUCACAUGGUGCAGCCCUACAUGCAUGUGAGCGAUAACCGUGAACUUGGUAU ((((....(((..........(((((((((((............)))--))))))))((((((.((((((((((........))).)))))))..)))))).)))...))))... ( -30.10, z-score = -2.14, R) >droGri2.scaffold_15245 14020543 113 - 18325388 UUAACUUACACUUAUUUGUAACAAUUUGAUGAUUUUGUUUCUCACUAU--UCAGGUCGUUAUCAGCACAUGCCACAACCAUACAAUCAUGUGAACGAUCAUCGUGAGCUUGGAGU (((((((((.......((.(((((..........)))))...))....--...(((((((.....((((((...............)))))))))))))...))))).))))... ( -17.46, z-score = 0.99, R) >consensus ______UAGACAG_CCCACUAACUU___CCGAUUUUGUUUCUCACCCU__AAAGGUCGCUAUCAUCACAUGCCGCAGCCCUACAGACAUGUGAGCGAUCAGCGUGAGCUUGGAAU .....................................................(((((((.....((((((...............)))))))))))))................ ( -8.99 = -9.61 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:49 2011