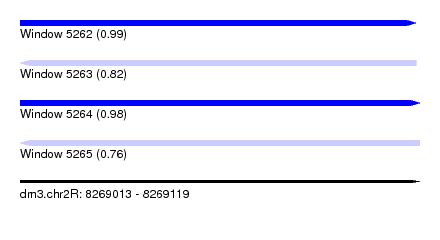
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,269,013 – 8,269,119 |
| Length | 106 |
| Max. P | 0.989665 |

| Location | 8,269,013 – 8,269,118 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.25 |
| Shannon entropy | 0.44909 |
| G+C content | 0.37646 |
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -12.88 |
| Energy contribution | -14.14 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
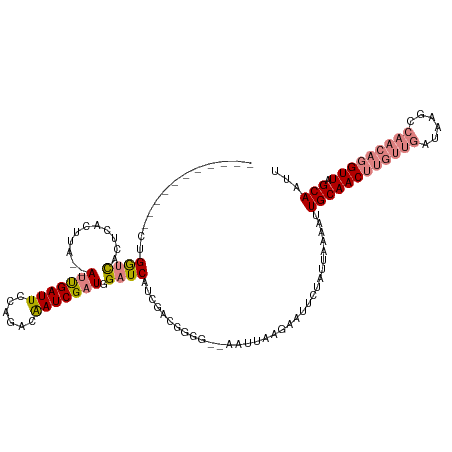
>dm3.chr2R 8269013 105 + 21146708 -----------CUGGUCACUCACUUA--AUUGAUUCCAGGCAAUCGAUGGAUCAUCGACGGGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUG -----------...............--.((((((((......((((((...))))))...))--)))))).............((((((((((((((......))))))))).))))). ( -28.20, z-score = -3.15, R) >droPer1.super_2 7681953 107 + 9036312 UAGAAUCAUCACGGAUUACUCACUUAUAAGUGAUUCCGUUCGAUCGAUGGAUCGCUCGUAGGAAUGUGUAUGUAUUCUAUUAAAACUGCAAC-------------AACAGGUUAGCAUUU (((((((((((((((....)).((((..((((((.(((((.....)))))))))))..))))..)))).))).)))))).......((((((-------------.....))).)))... ( -27.40, z-score = -2.10, R) >dp4.chr3 7484397 107 + 19779522 UAGAAUCAUCACGGAUUACUCACUUAUAAGUGAUUCCGUACGAUCGAUGGAUCGCUUGCAGGAAUGUGUAUGUAUUCUAUUAAAACUGCAAC-------------AACAGGUUAGCAUUU (((((((((((((......(((((....)))))((((((((((((....)))))..))).)))))))).))).)))))).......((((((-------------.....))).)))... ( -26.70, z-score = -1.95, R) >droEre2.scaffold_4845 5065316 107 - 22589142 -----------CCGGGCCCUCAAUUA--AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGGUCGAUUAAGAAUUCUAUUAAAACUGCAACUUGUUGAUAAGUCAACAGGUUAGCAAUU -----------............(((--(((((((((......((((((...)))))).))))))))))))...............(((((((((((((....)))))))))).)))... ( -30.60, z-score = -2.79, R) >droYak2.chr2R 12461873 105 - 21139217 -----------CCGGCCACUCAAUUA--AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGG--AAUUAAGAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUU -----------...............--.((((((((......((((((...))))))...))--))))))............(((((((((((((((......))))))))).)))))) ( -28.40, z-score = -3.10, R) >droSec1.super_1 5813737 105 + 14215200 -----------CUGGUCACUCACUUA--AUCGAUUCCAGACAAUCGAUGGAUCAUCGAAGUGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUG -----------((((....((.....--...))..))))....((((((...))))))(((((--(((.....))))))))...((((((((((((((......))))))))).))))). ( -29.40, z-score = -3.95, R) >droSim1.chr2R 6739070 105 + 19596830 -----------CUGGUCACUCACUUA--AUCGAUUCCAGACAAUCGAUGGAUCAUCGAAGUGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUG -----------((((....((.....--...))..))))....((((((...))))))(((((--(((.....))))))))...((((((((((((((......))))))))).))))). ( -29.40, z-score = -3.95, R) >consensus ___________CUGGUCACUCACUUA__AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGG__AAUUAAGAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUU .............((((...........(((((((......))))))).)))).................................((((((((((((......))))))))).)))... (-12.88 = -14.14 + 1.26)
| Location | 8,269,013 – 8,269,118 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Shannon entropy | 0.44909 |
| G+C content | 0.37646 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.61 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8269013 105 - 21146708 CAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCCCGUCGAUGAUCCAUCGAUUGCCUGGAAUCAAU--UAAGUGAGUGACCAG----------- .(((((.(((.((((((....)))))).)))))))).......((((.....)))--)...(((((((...)))))))...((((..(((..--....)))....))))----------- ( -21.70, z-score = -1.44, R) >droPer1.super_2 7681953 107 - 9036312 AAAUGCUAACCUGUU-------------GUUGCAGUUUUAAUAGAAUACAUACACAUUCCUACGAGCGAUCCAUCGAUCGAACGGAAUCACUUAUAAGUGAGUAAUCCGUGAUGAUUCUA ..........((((.-------------...))))......((((((.(((.(((........(..(((((....)))))..)(((.(((((....)))))....))))))))))))))) ( -25.60, z-score = -2.27, R) >dp4.chr3 7484397 107 - 19779522 AAAUGCUAACCUGUU-------------GUUGCAGUUUUAAUAGAAUACAUACACAUUCCUGCAAGCGAUCCAUCGAUCGUACGGAAUCACUUAUAAGUGAGUAAUCCGUGAUGAUUCUA ..........((((.-------------...))))......((((((.(((.(((..........((((((....))))))..(((.(((((....)))))....))))))))))))))) ( -26.70, z-score = -2.55, R) >droEre2.scaffold_4845 5065316 107 + 22589142 AAUUGCUAACCUGUUGACUUAUCAACAAGUUGCAGUUUUAAUAGAAUUCUUAAUCGACCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU--UAAUUGAGGGCCCGG----------- ((((((.(((.((((((....)))))).)))))))))...............((((((...))))))(..((.(((((((..((....))..--)))))))))..)...----------- ( -26.10, z-score = -1.75, R) >droYak2.chr2R 12461873 105 + 21139217 AAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUCUUAAUU--CCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU--UAAUUGAGUGGCCGG----------- ((((((.(((.((((((....)))))).)))))))))......((((.....)))--)...(((((((...)))))))...((((..((((.--...))))....))))----------- ( -23.20, z-score = -1.30, R) >droSec1.super_1 5813737 105 - 14215200 CAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCACUUCGAUGAUCCAUCGAUUGUCUGGAAUCGAU--UAAGUGAGUGACCAG----------- .(((((.(((.((((((....)))))).)))))))).............(..(((--(((..((((((...)))))).....))))))..).--...............----------- ( -21.90, z-score = -1.18, R) >droSim1.chr2R 6739070 105 - 19596830 CAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCACUUCGAUGAUCCAUCGAUUGUCUGGAAUCGAU--UAAGUGAGUGACCAG----------- .(((((.(((.((((((....)))))).)))))))).............(..(((--(((..((((((...)))))).....))))))..).--...............----------- ( -21.90, z-score = -1.18, R) >consensus AAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUCUUAAUU__CCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU__UAAGUGAGUGACCAG___________ .(((((.(((.((((((....)))))).))))))))............................(((((((....))))))).((..(((((....)))))....))............. (-13.26 = -14.61 + 1.35)

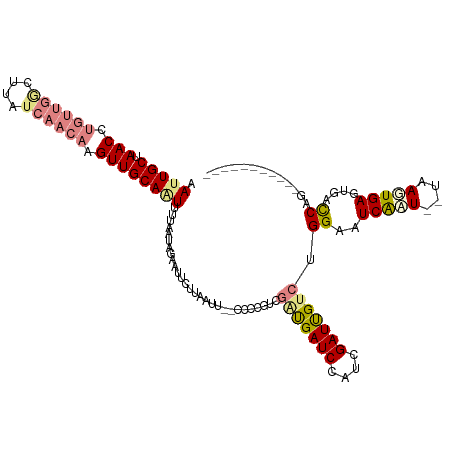
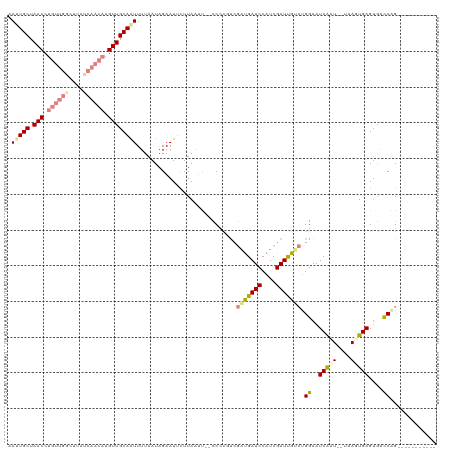
| Location | 8,269,013 – 8,269,119 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Shannon entropy | 0.45339 |
| G+C content | 0.37930 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -12.88 |
| Energy contribution | -14.14 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8269013 106 + 21146708 ----------CUGGUCACUCACUUA--AUUGAUUCCAGGCAAUCGAUGGAUCAUCGACGGGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUGC ----------...............--.((((((((......((((((...))))))...))--)))))).............((((((((((((((......))))))))).))))).. ( -28.20, z-score = -2.80, R) >droPer1.super_2 7681954 107 + 9036312 AGAAUCAUCACGGAUUACUCACUUAUAAGUGAUUCCGUUCGAUCGAUGGAUCGCUCGUAGGAAUGUGUAUGUAUUCUAUUAAAACUGCAAC-------------AACAGGUUAGCAUUUA ((((((((((((((....)).((((..((((((.(((((.....)))))))))))..))))..)))).))).)))))........((((((-------------.....))).))).... ( -26.20, z-score = -1.62, R) >dp4.chr3 7484398 107 + 19779522 AGAAUCAUCACGGAUUACUCACUUAUAAGUGAUUCCGUACGAUCGAUGGAUCGCUUGCAGGAAUGUGUAUGUAUUCUAUUAAAACUGCAAC-------------AACAGGUUAGCAUUUA ((((((((((((......(((((....)))))((((((((((((....)))))..))).)))))))).))).)))))........((((((-------------.....))).))).... ( -25.50, z-score = -1.46, R) >droEre2.scaffold_4845 5065316 108 - 22589142 ----------CCGGGCCCUCAAUUA--AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGGUCGAUUAAGAAUUCUAUUAAAACUGCAACUUGUUGAUAAGUCAACAGGUUAGCAAUUC ----------............(((--(((((((((......((((((...)))))).))))))))))))...............(((((((((((((....)))))))))).))).... ( -30.60, z-score = -2.77, R) >droYak2.chr2R 12461873 106 - 21139217 ----------CCGGCCACUCAAUUA--AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGG--AAUUAAGAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUUU ----------...............--.((((((((......((((((...))))))...))--))))))...........((((((((((((((((......))))))))).))))))) ( -29.30, z-score = -3.36, R) >droSec1.super_1 5813737 106 + 14215200 ----------CUGGUCACUCACUUA--AUCGAUUCCAGACAAUCGAUGGAUCAUCGAAGUGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUGC ----------((((....((.....--...))..))))....((((((...))))))(((((--(((.....))))))))...((((((((((((((......))))))))).))))).. ( -29.40, z-score = -3.63, R) >droSim1.chr2R 6739070 106 + 19596830 ----------CUGGUCACUCACUUA--AUCGAUUCCAGACAAUCGAUGGAUCAUCGAAGUGG--AAUUAAAAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUGC ----------((((....((.....--...))..))))....((((((...))))))(((((--(((.....))))))))...((((((((((((((......))))))))).))))).. ( -29.40, z-score = -3.63, R) >consensus __________CUGGUCACUCACUUA__AUUGAUUCCAGACAAUCGAUGGAUCAUCGACGGGG__AAUUAAGAAUUCUAUUAAAAUUGCAACUUGUUGAUAAGCCAACAGGUUAGCAAUUC ............((((...........(((((((......))))))).)))).................................((((((((((((......))))))))).))).... (-12.88 = -14.14 + 1.26)

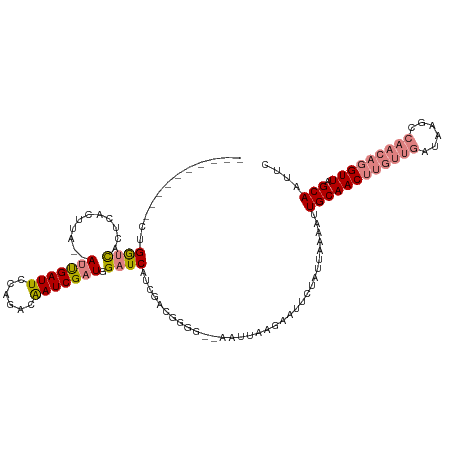
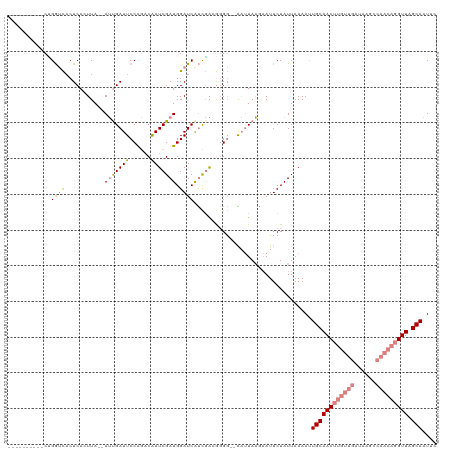
| Location | 8,269,013 – 8,269,119 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.19 |
| Shannon entropy | 0.45339 |
| G+C content | 0.37930 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.61 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8269013 106 - 21146708 GCAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCCCGUCGAUGAUCCAUCGAUUGCCUGGAAUCAAU--UAAGUGAGUGACCAG---------- (((((((.(((.((((((....)))))).)))))))....................--....(((((((...))))))))))((((..(((..--....)))....))))---------- ( -23.00, z-score = -1.58, R) >droPer1.super_2 7681954 107 - 9036312 UAAAUGCUAACCUGUU-------------GUUGCAGUUUUAAUAGAAUACAUACACAUUCCUACGAGCGAUCCAUCGAUCGAACGGAAUCACUUAUAAGUGAGUAAUCCGUGAUGAUUCU ...........((((.-------------...)))).......(((((.(((.(((........(..(((((....)))))..)(((.(((((....)))))....)))))))))))))) ( -24.20, z-score = -1.89, R) >dp4.chr3 7484398 107 - 19779522 UAAAUGCUAACCUGUU-------------GUUGCAGUUUUAAUAGAAUACAUACACAUUCCUGCAAGCGAUCCAUCGAUCGUACGGAAUCACUUAUAAGUGAGUAAUCCGUGAUGAUUCU ...........((((.-------------...)))).......(((((.(((.(((..........((((((....))))))..(((.(((((....)))))....)))))))))))))) ( -25.30, z-score = -2.17, R) >droEre2.scaffold_4845 5065316 108 + 22589142 GAAUUGCUAACCUGUUGACUUAUCAACAAGUUGCAGUUUUAAUAGAAUUCUUAAUCGACCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU--UAAUUGAGGGCCCGG---------- (((((((.(((.((((((....)))))).))))))))))..............((((((...))))))(..((.(((((((..((....))..--)))))))))..)...---------- ( -27.40, z-score = -1.97, R) >droYak2.chr2R 12461873 106 + 21139217 AAAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUCUUAAUU--CCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU--UAAUUGAGUGGCCGG---------- (((((((.(((.((((((....)))))).)))))))))).....((((.....)))--)...(((((((...)))))))...((((..((((.--...))))....))))---------- ( -24.10, z-score = -1.59, R) >droSec1.super_1 5813737 106 - 14215200 GCAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCACUUCGAUGAUCCAUCGAUUGUCUGGAAUCGAU--UAAGUGAGUGACCAG---------- ..(((((.(((.((((((....)))))).)))))))).............(..(((--(((..((((((...)))))).....))))))..).--...............---------- ( -21.90, z-score = -0.96, R) >droSim1.chr2R 6739070 106 - 19596830 GCAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUUUUAAUU--CCACUUCGAUGAUCCAUCGAUUGUCUGGAAUCGAU--UAAGUGAGUGACCAG---------- ..(((((.(((.((((((....)))))).)))))))).............(..(((--(((..((((((...)))))).....))))))..).--...............---------- ( -21.90, z-score = -0.96, R) >consensus GAAUUGCUAACCUGUUGGCUUAUCAACAAGUUGCAAUUUUAAUAGAAUUCUUAAUU__CCCCGUCGAUGAUCCAUCGAUUGUCUGGAAUCAAU__UAAGUGAGUGACCAG__________ ..(((((.(((.((((((....)))))).))))))))............................(((((((....))))))).((..(((((....)))))....))............ (-13.26 = -14.61 + 1.35)

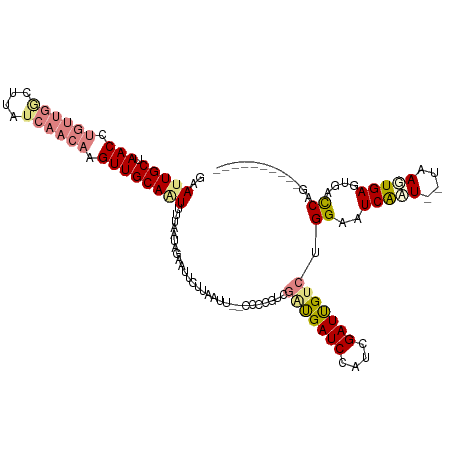
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:48 2011