
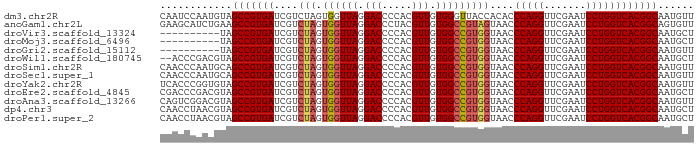
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,224,225 – 8,224,314 |
| Length | 89 |
| Max. P | 0.982290 |

| Location | 8,224,225 – 8,224,314 |
|---|---|
| Length | 89 |
| Sequences | 13 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.23199 |
| G+C content | 0.56451 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
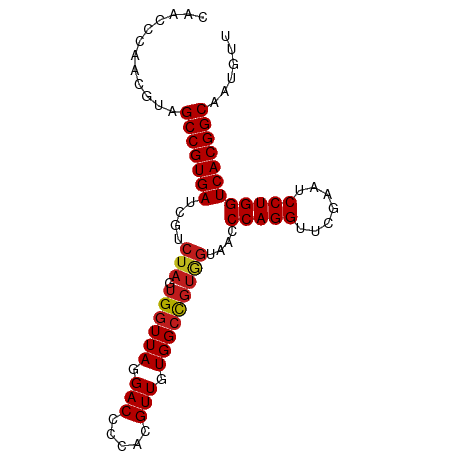
>dm3.chr2R 8224225 89 + 21146708 CAAUCCAAUGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGGUUACCACACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ......(((((.(((((((.......(((((...((((((.....).))))))))))..(((((.......)))))))))))).))))) ( -34.20, z-score = -2.83, R) >anoGam1.chr2L 29241518 89 + 48795086 GAAGCAUCUGAAGCCGUGAUCGUCUAGUGGUUAGGACCCUACGUUGUGGCCGUAGUAACCCAGGUUCGAAUCCUGGUCACGGCAGUGUU ............(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.30, z-score = -1.73, R) >droVir3.scaffold_13324 310157 79 - 2960039 ----------UAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU ----------..(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -1.77, R) >droMoj3.scaffold_6496 6172209 79 - 26866924 ----------UAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU ----------..(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -1.77, R) >droGri2.scaffold_15112 271471 79 - 5172618 ----------UAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ----------..(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -2.06, R) >droWil1.scaffold_180745 2384232 87 - 2843958 --ACCCGACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU --....(.(((.(((((((....(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).)))). ( -30.90, z-score = -0.86, R) >droSim1.chr2R 6692958 89 + 19596830 CAACCCAAUGCAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ............(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -0.99, R) >droSec1.super_1 5770148 89 + 14215200 CAACCCAAUGCAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ............(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -0.99, R) >droYak2.chr2R 8728696 89 + 21139217 UCACCCGGUGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ............(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... ( -30.00, z-score = -0.45, R) >droEre2.scaffold_4845 5021045 89 - 22589142 CGACCCGACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU ......(.(((.(((((((....(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).)))). ( -30.90, z-score = -0.49, R) >droAna3.scaffold_13266 5516347 89 + 19884421 CAGUCGGACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ......(((((.(((((((....(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))))) ( -33.20, z-score = -1.55, R) >dp4.chr3 2800258 89 + 19779522 CAACCUAACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU ........(((.(((((((....(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))).. ( -30.70, z-score = -1.20, R) >droPer1.super_2 2990271 89 + 9036312 CAACCUAACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCU ........(((.(((((((....(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))).. ( -30.70, z-score = -1.20, R) >consensus CAACCCAACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUU ............(((((((....(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))...... (-28.78 = -29.03 + 0.25)
| Location | 8,224,225 – 8,224,314 |
|---|---|
| Length | 89 |
| Sequences | 13 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.23199 |
| G+C content | 0.56451 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -29.91 |
| Energy contribution | -30.24 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
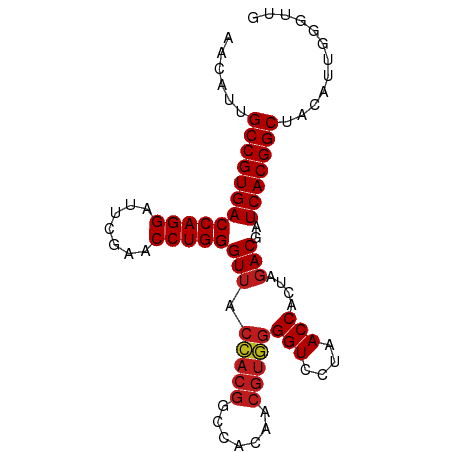
>dm3.chr2R 8224225 89 - 21146708 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUGUGGUAACCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUGGAUUG ......((((((((((((.......)))))..(((((..(((((...)))))......))))).......)))))))............ ( -32.80, z-score = -2.32, R) >anoGam1.chr2L 29241518 89 - 48795086 AACACUGCCGUGACCAGGAUUCGAACCUGGGUUACUACGGCCACAACGUAGGGUCCUAACCACUAGACGAUCACGGCUUCAGAUGCUUC ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ ( -29.70, z-score = -2.56, R) >droVir3.scaffold_13324 310157 79 + 2960039 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUA---------- ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))..---------- ( -31.70, z-score = -2.95, R) >droMoj3.scaffold_6496 6172209 79 + 26866924 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUA---------- ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))..---------- ( -31.70, z-score = -2.95, R) >droGri2.scaffold_15112 271471 79 + 5172618 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUA---------- ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))..---------- ( -31.70, z-score = -3.33, R) >droWil1.scaffold_180745 2384232 87 + 2843958 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCGGGU-- ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))..........-- ( -31.70, z-score = -1.18, R) >droSim1.chr2R 6692958 89 - 19596830 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUGGGUUG ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ ( -31.70, z-score = -1.53, R) >droSec1.super_1 5770148 89 - 14215200 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUGGGUUG ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ ( -31.70, z-score = -1.53, R) >droYak2.chr2R 8728696 89 - 21139217 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACACCGGGUGA ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))..(((...))). ( -31.90, z-score = -1.39, R) >droEre2.scaffold_4845 5021045 89 + 22589142 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCGGGUCG ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ ( -31.70, z-score = -0.95, R) >droAna3.scaffold_13266 5516347 89 - 19884421 AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCCGACUG ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ ( -31.70, z-score = -2.08, R) >dp4.chr3 2800258 89 - 19779522 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUUAGGUUG (((...((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))...)))...... ( -32.50, z-score = -1.94, R) >droPer1.super_2 2990271 89 - 9036312 AGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUUAGGUUG (((...((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))...)))...... ( -32.50, z-score = -1.94, R) >consensus AACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUGGGUUG ......((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))............ (-29.91 = -30.24 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:41 2011