
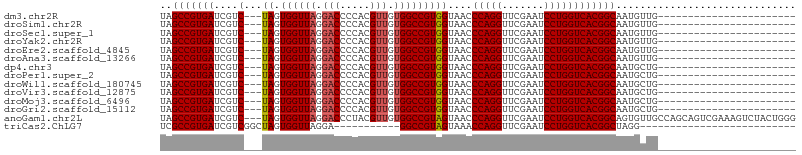
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,223,920 – 8,224,000 |
| Length | 80 |
| Max. P | 0.997135 |

| Location | 8,223,920 – 8,224,000 |
|---|---|
| Length | 80 |
| Sequences | 14 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Shannon entropy | 0.17533 |
| G+C content | 0.56844 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.35 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
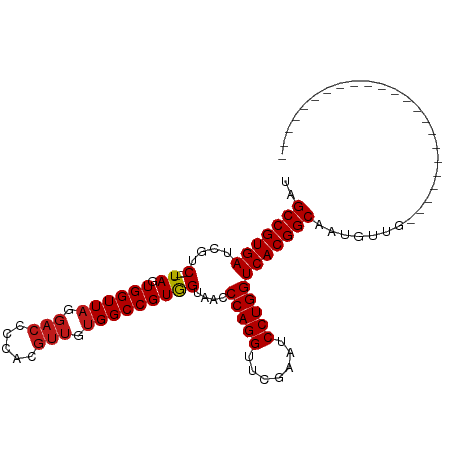
>dm3.chr2R 8223920 80 + 21146708 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >droSim1.chr2R 6692663 80 + 19596830 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >droSec1.super_1 5769853 80 + 14215200 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >droYak2.chr2R 12412764 80 - 21139217 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >droEre2.scaffold_4845 5020693 80 - 22589142 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >droAna3.scaffold_13266 5515682 80 + 19884421 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -2.03, R) >dp4.chr3 16981574 80 - 19779522 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >droPer1.super_2 6048362 80 - 9036312 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >droWil1.scaffold_180745 463085 80 + 2843958 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >droVir3.scaffold_12875 13998727 80 - 20611582 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >droMoj3.scaffold_6496 23396889 80 + 26866924 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >droGri2.scaffold_15112 3672713 80 - 5172618 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUG----------------------- ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......----------------------- ( -30.00, z-score = -1.74, R) >anoGam1.chr2L 19560199 103 - 48795086 UAGCCGUGAUCGUC---UAGUGGUUAGGACCCUACGUUGUGGCCGUAGUAACCCAGGUUCGAAUCCUGGUCACGGCAGUGUUGCCAGCAGUCGAAAGUCUACUGGG ..(((((((....(---((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).......((((.((.(....).)).)))). ( -40.60, z-score = -2.82, R) >triCas2.ChLG7 15875478 69 - 17478683 UCGCCGUGAUCGUCGGCUAGUGGUUAGGA-----------GGCCGUAGUAAACCAGGUUCGAAUCCUGGUCACGGCUAGG-------------------------- ..(((((((......((((.(((((....-----------)))))))))...(((((.......))))))))))))....-------------------------- ( -27.20, z-score = -2.33, R) >consensus UAGCCGUGAUCGUC___UAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUG_______________________ ..(((((((............(((((.(((.....))).)))))........(((((.......)))))))))))).............................. (-28.12 = -28.35 + 0.23)
| Location | 8,223,920 – 8,224,000 |
|---|---|
| Length | 80 |
| Sequences | 14 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Shannon entropy | 0.17533 |
| G+C content | 0.56844 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
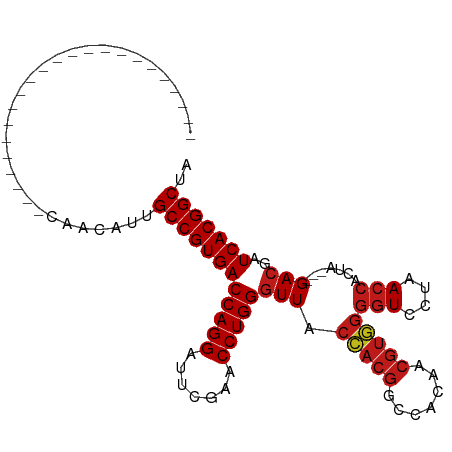
>dm3.chr2R 8223920 80 - 21146708 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >droSim1.chr2R 6692663 80 - 19596830 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >droSec1.super_1 5769853 80 - 14215200 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >droYak2.chr2R 12412764 80 + 21139217 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >droEre2.scaffold_4845 5020693 80 + 22589142 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >droAna3.scaffold_13266 5515682 80 - 19884421 -----------------------CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.34, R) >dp4.chr3 16981574 80 + 19779522 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >droPer1.super_2 6048362 80 + 9036312 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >droWil1.scaffold_180745 463085 80 - 2843958 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >droVir3.scaffold_12875 13998727 80 + 20611582 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >droMoj3.scaffold_6496 23396889 80 - 26866924 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >droGri2.scaffold_15112 3672713 80 + 5172618 -----------------------CAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA---GACGAUCACGGCUA -----------------------.......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -31.70, z-score = -3.00, R) >anoGam1.chr2L 19560199 103 + 48795086 CCCAGUAGACUUUCGACUGCUGGCAACACUGCCGUGACCAGGAUUCGAACCUGGGUUACUACGGCCACAACGUAGGGUCCUAACCACUA---GACGAUCACGGCUA .(((((((........))))))).......((((((((((((.......)))))(((.(((((.......)))))(((....)))....---)))..))))))).. ( -39.00, z-score = -3.47, R) >triCas2.ChLG7 15875478 69 + 17478683 --------------------------CCUAGCCGUGACCAGGAUUCGAACCUGGUUUACUACGGCC-----------UCCUAACCACUAGCCGACGAUCACGGCGA --------------------------....((((((((((((.......))))).......((((.-----------............))))....))))))).. ( -25.42, z-score = -3.76, R) >consensus _______________________CAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUA___GACGAUCACGGCUA ..............................((((((((((((.......)))))(((.(((((.......)))))(((....))).......)))..))))))).. (-28.92 = -29.36 + 0.45)

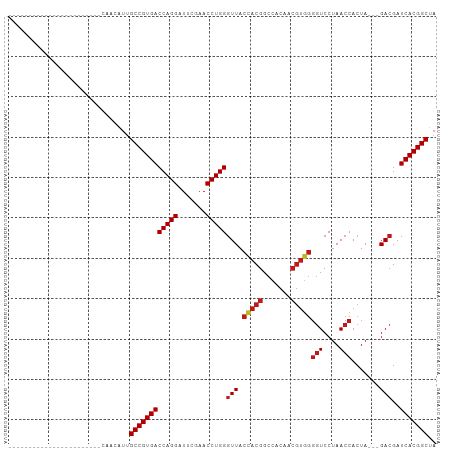
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:39 2011