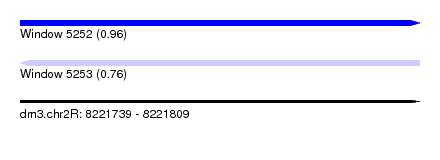
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,221,739 – 8,221,809 |
| Length | 70 |
| Max. P | 0.956252 |

| Location | 8,221,739 – 8,221,809 |
|---|---|
| Length | 70 |
| Sequences | 14 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 96.40 |
| Shannon entropy | 0.08936 |
| G+C content | 0.59658 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -23.63 |
| Energy contribution | -24.01 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8221739 70 + 21146708 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droSim1.chr2R 6690466 70 + 19596830 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droSec1.super_1 5767637 70 + 14215200 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droYak2.chr2R 12410438 70 - 21139217 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droEre2.scaffold_4845 5018593 70 - 22589142 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droAna3.scaffold_13266 5513618 70 + 19884421 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >dp4.chr3 16979181 70 - 19779522 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droPer1.super_2 6045958 70 - 9036312 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droWil1.scaffold_180745 460670 70 + 2843958 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droVir3.scaffold_13324 2649809 70 + 2960039 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droMoj3.scaffold_6496 642387 70 - 26866924 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >droGri2.scaffold_15112 4901074 70 + 5172618 GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -27.30, z-score = -2.33, R) >anoGam1.chr2L 19553485 70 - 48795086 GCCGUGACCAGGAUUCGAACCUGGGUUACUACGGCCACAACGUAGGGUCCUAACCACUAG---ACGAUCACGG .(((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))) ( -25.30, z-score = -2.45, R) >triCas2.ChLG7 1603140 62 + 17478683 GCCGUGACCAGGAUUCGAACCUGGUUUACUACGGCC-----------UCCUAACCACUAGCCGACGAUCACGG .(((((((((((.......))))).......((((.-----------............))))....)))))) ( -20.32, z-score = -3.06, R) >consensus GCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG___ACGAUCACGG .(((((((((((.......)))))....(((((.......)))))(((....)))............)))))) (-23.63 = -24.01 + 0.38)

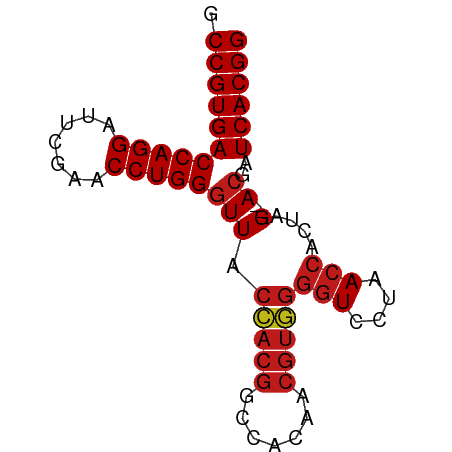
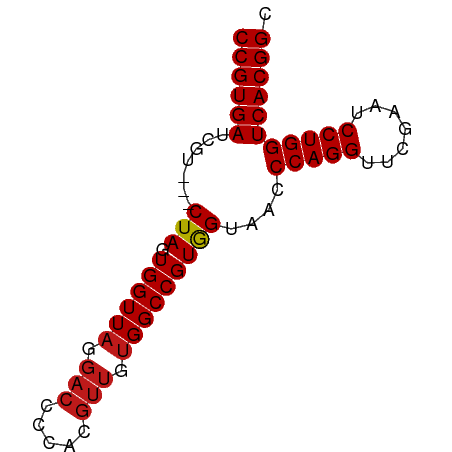
| Location | 8,221,739 – 8,221,809 |
|---|---|
| Length | 70 |
| Sequences | 14 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 96.40 |
| Shannon entropy | 0.08936 |
| G+C content | 0.59658 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8221739 70 - 21146708 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droSim1.chr2R 6690466 70 - 19596830 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droSec1.super_1 5767637 70 - 14215200 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droYak2.chr2R 12410438 70 + 21139217 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droEre2.scaffold_4845 5018593 70 + 22589142 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droAna3.scaffold_13266 5513618 70 - 19884421 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >dp4.chr3 16979181 70 + 19779522 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droPer1.super_2 6045958 70 + 9036312 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droWil1.scaffold_180745 460670 70 - 2843958 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droVir3.scaffold_13324 2649809 70 - 2960039 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droMoj3.scaffold_6496 642387 70 + 26866924 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >droGri2.scaffold_15112 4901074 70 - 5172618 CCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.10, z-score = -1.00, R) >anoGam1.chr2L 19553485 70 + 48795086 CCGUGAUCGU---CUAGUGGUUAGGACCCUACGUUGUGGCCGUAGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). ( -25.40, z-score = -1.78, R) >triCas2.ChLG7 1603140 62 - 17478683 CCGUGAUCGUCGGCUAGUGGUUAGGA-----------GGCCGUAGUAAACCAGGUUCGAAUCCUGGUCACGGC ((((((......((((.(((((....-----------)))))))))...(((((.......))))))))))). ( -22.70, z-score = -2.09, R) >consensus CCGUGAUCGU___CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGC ((((((.......(((.((((((.(((.....))).)))))))))....(((((.......))))))))))). (-24.43 = -24.54 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:38 2011