| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,200,914 – 8,201,048 |
| Length | 134 |
| Max. P | 0.995115 |

| Location | 8,200,914 – 8,201,048 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 70.36 |
| Shannon entropy | 0.57334 |
| G+C content | 0.41095 |
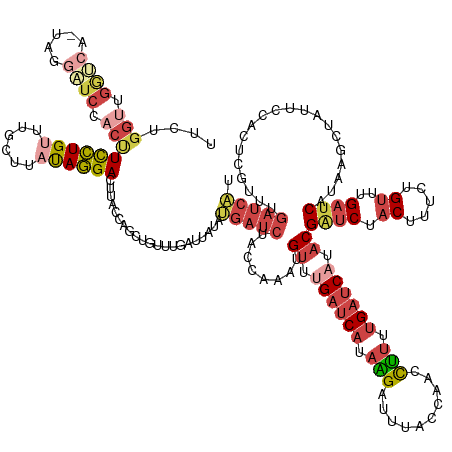
| Mean single sequence MFE | -35.79 |
| Consensus MFE | -15.61 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
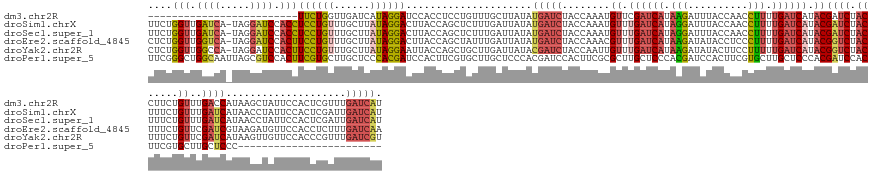
>dm3.chr2R 8200914 134 - 21146708 -------------------------UUCUGGUUGAUCAUAGGAUCCACCUCCUGUUUGCUUAUAUGAUCUACCAAAUGUUCGAUCAUAAGAUUUACCAACCUUUUGAUCAUACGAUCUACCUUCUGUUUGACCAUAAGCUAUUCCACUCGUUUGAUCAU -------------------------...((((.((((((((((......)))))).......(((((((.((.....))..))))))).)))).))))......((((((.((((..........(((((....)))))........)))).)))))). ( -30.27, z-score = -3.21, R) >droSim1.chrX 2719273 158 + 17042790 UUCUGGUUGAUCA-UAGGAUCCACCUCCUGUUUGCUUAUAGGACUUACCAGCUCUUUGAUUAUAUGAUCUACCAAAUGUUUGAUCAUAGGAUUUACCAACCUUUUGAUCAUACGAUCUACUUUCUGUUUGAUCAUAACCUAUUCCACUCGAUUGAUCAU ...((((..((((-((((((((...((((((......))))))...................(((((((.((.....))..)))))))))))............((((((.((((.......)).)).))))))...))))).......)))..)))). ( -34.81, z-score = -2.36, R) >droSec1.super_1 5749361 158 - 14215200 UUCUGGUUGAUCA-UAGGAUCCACCUCCUGUUUGCUUAUAGGACUUACCAGCUCUUUGAUUAUAUGAUCUACCAAAUGUUUGAUCAUAGGAUUUACCAACCUUUUGAUCAUACGAUCUACUUUCUGUUUGAUCAUAACCUAUUCCACUCGAUUGAUCAU ...((((..((((-((((((((...((((((......))))))...................(((((((.((.....))..)))))))))))............((((((.((((.......)).)).))))))...))))).......)))..)))). ( -34.81, z-score = -2.36, R) >droEre2.scaffold_4845 4995998 158 + 22589142 CUCUGGUUGGUCA-UAGGAUCCACUUCCUGUUUGCUUAUAGGACUUACCAGCUAUUUGAUUAUAUGAUCUACCAAACGUUUGAUCAUAAGAUAUACCUCCCUUUUGAUCAUACGGUCUACUUUCUGUUCGAUCGUAAGAUGUUCCACCUCUUUGAUCAA ...((((.(((((-((.((((....((((((......))))))((....))......)))).))))))).))))..(((.((((((.(((..........))).)))))).)))...............(((((.((((.(.....).))))))))).. ( -38.20, z-score = -2.29, R) >droYak2.chr2R 12391382 158 + 21139217 CUCUGGUUGGCCA-UAGGAUCCACUUCCUGUUUGCUUAUAGGAAUUACCAGCUGCUUGAUUAUACGAUCUACCAAUUGUUUGAUCAUAAGAUAUACUUCCUUUUUGAUCAUACGGUCUACUUUCUGUUCGAUCAUAAGUUGUUCCACCCGUUUGAUCGU ....((.(((..(-(((((......))))))..(((((((((((.((....((...((((((.(((((......))))).))))))..))...)).)))))....((((..((((........))))..))))))))))....))).)).......... ( -35.50, z-score = -1.42, R) >droPer1.super_5 4504831 135 + 6813705 UUCGGGCUGGCAAUUAGCGUCCACUUCGUGCUUGCUCCCACGAUCCACUUCGUGCUUGCUCCCACGAUCCACUUCGCGCUUGCUCCCACGAUCCACUUCGUGCUUGCUCCCACGAUCCACUUCGUGCUUGCUCCC------------------------ ...(((..(((((...(((......(((((...((...(((((......)))))...))...))))).......)))..))))))))((((......))))....((...(((((......)))))...))....------------------------ ( -41.12, z-score = -4.17, R) >consensus UUCUGGUUGGUCA_UAGGAUCCACUUCCUGUUUGCUUAUAGGACUUACCAGCUGUUUGAUUAUAUGAUCUACCAAAUGUUUGAUCAUAAGAUUUACCAACCUUUUGAUCAUACGAUCUACUUUCUGUUUGAUCAUAAGCUAUUCCACUCGUUUGAUCAU ....(((.((((.....)))).)))((((((......)))))).....................(((((........((.((((((.(((..........))).)))))).))((((.((.....))..))))....................))))). (-15.61 = -17.13 + 1.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:35 2011