| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,178,553 – 8,178,660 |
| Length | 107 |
| Max. P | 0.500000 |

| Location | 8,178,553 – 8,178,660 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.60773 |
| G+C content | 0.52676 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -11.30 |
| Energy contribution | -13.19 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
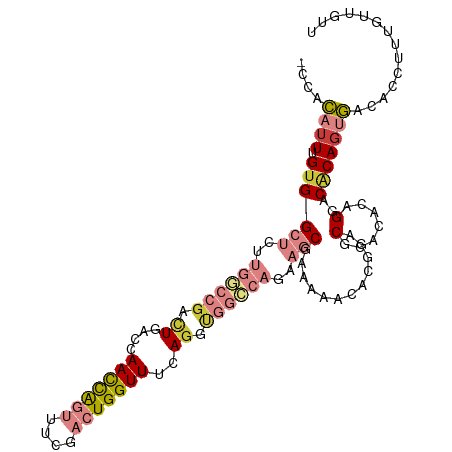
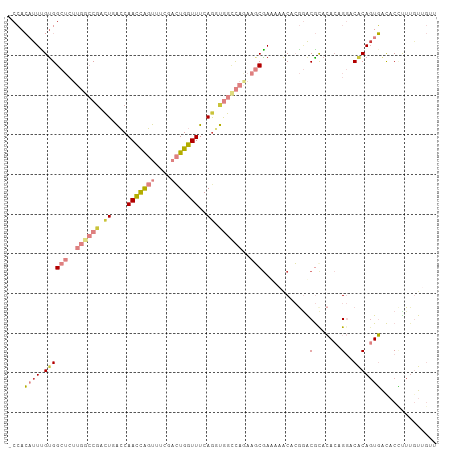
>dm3.chr2R 8178553 107 - 21146708 -CCACAUUCGUGGCUCUUGACCGACUGACCAACCAGUUUCGACUGGUUUCAGGUGGCCAGAAGCGAAAAACCCGGACGCACAGAGGACACAGUGACACCUUUGUUGUU -.(((....)))(((..((.(((.((((..(((((((....))))))))))).))).))..)))..........((((.(((((((...........))))))))))) ( -32.30, z-score = -0.70, R) >droPer1.super_2 6005339 97 + 9036312 GCAAACUUUGUGGC-----AACGUUUCGGCAAUUGCCGGAAAUUGGUUUCAGCCAGUUUUAGCCGAC--GCAGAGGCACAGACCGACCGCAGUUGUUCGCUUGU---- ((.(((.(((((((-----.......((((....))))(((((((((....))))))))).(((...--.....))).......).))))))..))).))....---- ( -31.60, z-score = -1.01, R) >dp4.chr3 16938711 97 + 19779522 GCAAACUUUGUGGC-----AACGUUUCGGCAAUUGCCGGAAAUUGGUUUCAGCCAGUUUUAGCCGAC--GCAGAGGCACAGACCGACCGCAGUUGUUCGCUUGU---- ((.(((.(((((((-----.......((((....))))(((((((((....))))))))).(((...--.....))).......).))))))..))).))....---- ( -31.60, z-score = -1.01, R) >droEre2.scaffold_4845 4973437 106 + 22589142 -CCACAUUUGUGGCUCUUGGCCGACUGACCAACCAGUUUCGACUGGUUUCAGGUGGGCAAAAGCAAAAACCACG-ACGCACACAGGACACAGUGGCACUUUUGUUGUU -..(((...((((((.(((.(((.((((..(((((((....))))))))))).))).))).))).....((((.-...(......).....)))))))...))).... ( -32.60, z-score = -0.72, R) >droYak2.chr2R 12368839 106 + 21139217 -CCACAUUUGUGGCUCUUGGCCCACUGACCAACCAGUUUCGACUGGUUUCAAGUGGCCAGAAGCGA-AAACCCGAGCGUACACAGGACACAGUGACACCUUUGUUGUU -.(((....)))(((..(((((.(((....(((((((....)))))))...))))))))..)))..-.......((((.(((.(((...........))).))))))) ( -32.20, z-score = -1.06, R) >droSec1.super_1 5726328 107 - 14215200 -CCACAUUCGUGGCUCUUGGCCGACUGACCAACCAGUUUAGACUGGUUUCAGUUGGCCAGAAGCGAUUAACACGGACGCACACAGGACACACUGACACCUUUCUUGUU -.....(((((((((..(((((((((((..(((((((....))))))))))))))))))..)))......))))))......(((......))).............. ( -40.30, z-score = -4.73, R) >droSim1.chr2R 6645459 107 - 19596830 -CCACAUUCGCGGCUCUUGGCCGACUCACCAACCAGUUUCGACUGGUUUCAGUUGGCCAGAAGCGAUUAACACGGACGCACACAGGACACAGUGACACCUUGAUGGGC -(((...((((...(((.((((((((....(((((((....)))))))..))))))))))).))))....((.((...(((..........)))...)).)).))).. ( -36.00, z-score = -2.35, R) >consensus _CCACAUUUGUGGCUCUUGGCCGACUGACCAACCAGUUUCGACUGGUUUCAGGUGGCCAGAAGCGAAAAACACGGACGCACACAGGACACAGUGACACCUUUGUUGUU ....((((.((((((.(((((((.((....(((((((....)))))))..)).))))))).)))............(.......)..))))))).............. (-11.30 = -13.19 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:33 2011