
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,176,784 – 8,176,883 |
| Length | 99 |
| Max. P | 0.858693 |

| Location | 8,176,784 – 8,176,883 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.55496 |
| G+C content | 0.48737 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
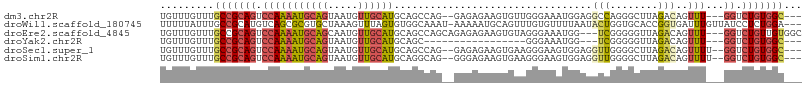
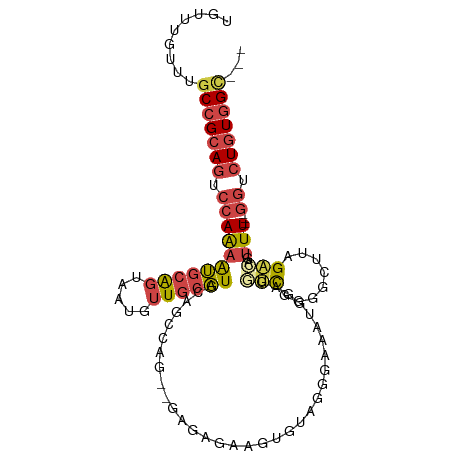
>dm3.chr2R 8176784 99 + 21146708 UGUUUGUUUGCCGCAGUCCAAAAUGCAGUAAUGUUGCAUGCAGCCAG--GAGAGAAGUGUUGGGAAAUGGAGGCCAGGGCUUAGACAGUUU---GGUCUGUGGC--- .........(((((((.(((((.(((((((....))).)))).....--.....((((.((((..........)))).))))......)))---)).)))))))--- ( -30.70, z-score = -0.87, R) >droWil1.scaffold_180745 669929 103 + 2843958 UUUUUAUUUGCCGCAUGUCAGCGCGUGCUAAAGUUUAGUGUGGCAAAU-AAAAAUGCAGUUUGUGUUUUAAUACUGGUGCACCGGUGAUUUGUUAUCCUCUGGA--- ((((((((((((((((...(((..........)))..)))))))))))-)))))((((.(..((((....)))).).))))((((.(((.....)))..)))).--- ( -31.80, z-score = -2.61, R) >droEre2.scaffold_4845 4971712 101 - 22589142 UGUUUGUUUGCCGCAGUCCAAAAUGCAGCAAUGUUGCAUGCAGCCAGCAGAGAGAAGUGUAGGGAAAUGG---UCGGGGGUUAGACAGUUU---GGUCUGUUGUGGC .........(((((((.(((...(((((((....))).)))).((.(((........))).))....)))---.)......(((((.....---.))))).)))))) ( -26.20, z-score = 0.54, R) >droYak2.chr2R 12367132 81 - 21139217 UGUUUGUUUGCCGCAGUCCAAAAUGCAGUAAUGUUGCAUGCAGC-----------------GGGAAAUGG---UCGGGGGUUAGACAGUUU---GGUCUGUGGC--- .........(((((((.(((((...((....(((((....))))-----------------).....))(---((........)))..)))---)).)))))))--- ( -26.40, z-score = -1.78, R) >droSec1.super_1 5724587 100 + 14215200 UGUUUGUUUGCCGCAGUCCAAAAUGCAGUAAUGUUGCAUGCAGCCAG--GAGAGAAGUGAAGGGAAGUGGAGGUUGGGGCUUAGACAGUUUU--GGUCUGUGGC--- .........(((((((.((((((((((....))).((.(.(((((..--......................))))).))).......)))))--)).)))))))--- ( -28.76, z-score = -0.94, R) >droSim1.chr2R 6643721 100 + 19596830 UGUUUGUUUGCCGCAGUCCAAAAUGCAGUAAUGUUGCAUGCAGGCAG--GGGAGAAGUGAAGGGAAGUGGAGGUUGGGGCUUAGACAGUUUU--GGUCUGUGGC--- .........(((((((.(((((((((((((....))).)))......--.................((..((((....))))..)).)))))--)).)))))))--- ( -27.80, z-score = -0.89, R) >consensus UGUUUGUUUGCCGCAGUCCAAAAUGCAGUAAUGUUGCAUGCAGCCAG__GAGAGAAGUGUAGGGAAAUGGAGGUCGGGGCUUAGACAGUUU___GGUCUGUGGC___ .........(((((((.((...((((((.....)))))).................................(((........)))........)).)))))))... (-14.12 = -14.82 + 0.70)

| Location | 8,176,784 – 8,176,883 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.55496 |
| G+C content | 0.48737 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.858693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
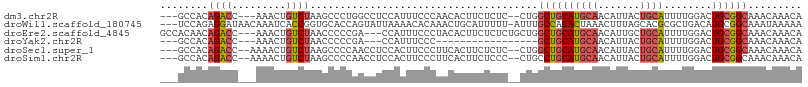
>dm3.chr2R 8176784 99 - 21146708 ---GCCACAGACC---AAACUGUCUAAGCCCUGGCCUCCAUUUCCCAACACUUCUCUC--CUGGCUGCAUGCAACAUUACUGCAUUUUGGACUGCGGCAAACAAACA ---(((.(((.((---(((........((...((((......................--..))))))(((((.......)))))))))).))).)))......... ( -23.16, z-score = -1.73, R) >droWil1.scaffold_180745 669929 103 - 2843958 ---UCCAGAGGAUAACAAAUCACCGGUGCACCAGUAUUAAAACACAAACUGCAUUUUU-AUUUGCCACACUAAACUUUAGCACGCGCUGACAUGCGGCAAAUAAAAA ---..(((..(((.....)))((.((....)).)).............)))..(((((-(((((((.((.......(((((....)))))..)).)))))))))))) ( -20.20, z-score = -1.46, R) >droEre2.scaffold_4845 4971712 101 + 22589142 GCCACAACAGACC---AAACUGUCUAACCCCCGA---CCAUUUCCCUACACUUCUCUCUGCUGGCUGCAUGCAACAUUGCUGCAUUUUGGACUGCGGCAAACAAACA (((.((..((((.---.....)))).....((((---.......((..((........))..)).((((.(((....)))))))..))))..)).)))......... ( -19.00, z-score = -0.91, R) >droYak2.chr2R 12367132 81 + 21139217 ---GCCACAGACC---AAACUGUCUAACCCCCGA---CCAUUUCCC-----------------GCUGCAUGCAACAUUACUGCAUUUUGGACUGCGGCAAACAAACA ---(((.(((.((---(((..(((........))---)........-----------------.....(((((.......)))))))))).))).)))......... ( -19.40, z-score = -3.12, R) >droSec1.super_1 5724587 100 - 14215200 ---GCCACAGACC--AAAACUGUCUAAGCCCCAACCUCCACUUCCCUUCACUUCUCUC--CUGGCUGCAUGCAACAUUACUGCAUUUUGGACUGCGGCAAACAAACA ---(((.(((.((--((((.(((...((((............................--..)))))))((((.......)))))))))).))).)))......... ( -21.11, z-score = -2.47, R) >droSim1.chr2R 6643721 100 - 19596830 ---GCCACAGACC--AAAACUGUCUAAGCCCCAACCUCCACUUCCCUUCACUUCUCCC--CUGCCUGCAUGCAACAUUACUGCAUUUUGGACUGCGGCAAACAAACA ---(((.(((.((--((((.......................................--.((....))((((.......)))))))))).))).)))......... ( -17.70, z-score = -1.97, R) >consensus ___GCCACAGACC___AAACUGUCUAAGCCCCAACCUCCAUUUCCCUACACUUCUCUC__CUGGCUGCAUGCAACAUUACUGCAUUUUGGACUGCGGCAAACAAACA ........((((.........))))......................................((((((((((.......))))........))))))......... ( -8.06 = -8.53 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:32 2011