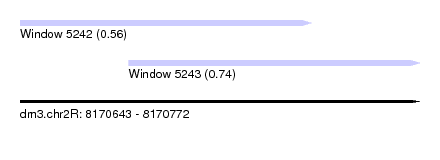
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,170,643 – 8,170,772 |
| Length | 129 |
| Max. P | 0.740382 |

| Location | 8,170,643 – 8,170,737 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.38 |
| Shannon entropy | 0.58215 |
| G+C content | 0.41562 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
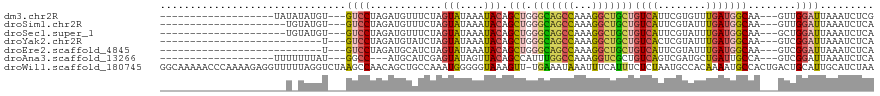
>dm3.chr2R 8170643 94 + 21146708 -------------------UAUAUAUGU---GUCCUAGAUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUGUUUGAUGGCAA---GUUGGAUUAAAUCUCG -------------------....(((.(---((.(((((....))))).))).)))(((((.(((((((...)))))))(((((.((....))))))).)---))))............ ( -26.40, z-score = -1.88, R) >droSim1.chr2R 6637180 92 + 19596830 ---------------------UGUAUGU---GUCCUAGAUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA---GUUGGAUUAAAUCUCA ---------------------.((((.(---((.(((((....))))).))).))))((((.(((((((...)))))))(((((.((....))))))).)---)))............. ( -26.60, z-score = -1.89, R) >droSec1.super_1 5718552 92 + 14215200 ---------------------UGUAUGU---GUCCUAGAUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA---GCUGGAUUAAAUCUCA ---------------------.((((.(---((.(((((....))))).))).))))((((.(((((((...)))))))(((((.((....))))))).)---)))............. ( -29.00, z-score = -2.31, R) >droYak2.chr2R 12361111 86 - 21139217 ---------------------------U---GUCCUAGAUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCACUCGUAUUUGAUGGCAA---GUCGGAUUAAAUCUCA ---------------------------(---((((((((....)))))..(((((((((.(((((((((...)))))))..)))).)))))))..)))).---................ ( -22.40, z-score = -0.91, R) >droEre2.scaffold_4845 4965773 86 - 22589142 ---------------------------U---GUCCUAGAUGCAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA---GUCGGAUUAAAUCUCA ---------------------------.---((((....(((..(((((.........))))))))......(((((((((((........))))))).)---)))))))......... ( -21.80, z-score = -0.58, R) >droAna3.scaffold_13266 5462674 91 + 19884421 -------------------UUUUUUUAU---GGCC---AUGCAUCGAGUAUAGUUACAGCCAUUUGGCCAAAGGUCGCUGUCAGUCGAUGCUGAUUGCCA---GUCGGAUUAAAUCUCA -------------------........(---(((.---..(((((((.((((((.((.(((....))).....)).))))).).))))))).....))))---................ ( -26.00, z-score = -1.79, R) >droWil1.scaffold_180745 661453 118 + 2843958 GGCAAAAACCCAAAAGAGGUUUUUAGGUCUAAGCCAACAGCUGCCAAAUGGGGGUAAAGUU-UGAAAUAAAUUUCAUUUCUCUAAUGCCACAAAAUGCCACUGACUGCAUUGCAUCUAA ((((...((((......(((((........)))))........((....))))))..((..-(((((....)))))...))....))))....(((((........)))))........ ( -22.90, z-score = 0.87, R) >consensus _____________________U_UAUGU___GUCCUAGAUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA___GUUGGAUUAAAUCUCA ...............................((((............(((....))).(((.(((((((...)))))))((((........)))))))........))))......... (-11.06 = -11.46 + 0.40)
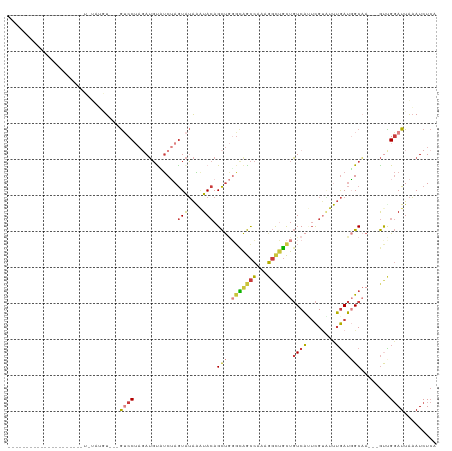
| Location | 8,170,678 – 8,170,772 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.21 |
| Shannon entropy | 0.56427 |
| G+C content | 0.47965 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
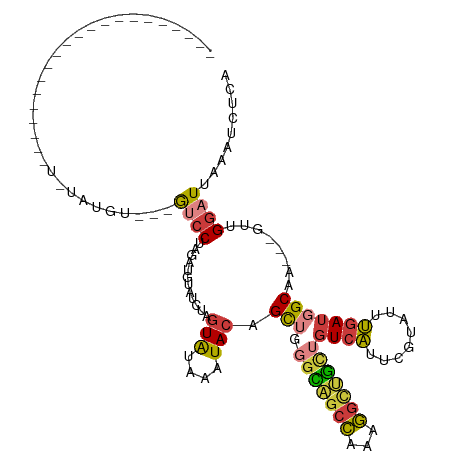
>dm3.chr2R 8170678 94 + 21146708 -------------------------AGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUGUUUGAUGGCAA-GUUGGAUUAAAUCUCGAUGGGGGAUAAAUUGUGGACGGUGGCCGGCAGCCA -------------------------.....(((.(((...(((..(((((......(((..((.....-))..)))...(((((.....)))))........)))))..)))))).))). ( -32.70, z-score = -1.09, R) >droSim1.chr2R 6637213 94 + 19596830 -------------------------AGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA-GUUGGAUUAAAUCUCAAUGGGGGAUAAAUUGUGUUUGGUGGCCGGCAGCCA -------------------------.(((((.((.(((((.((((((((((........)))))))).-..........(((((.....))))).....)).))))))).)))))..... ( -33.90, z-score = -1.88, R) >droSec1.super_1 5718585 94 + 14215200 -------------------------AGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA-GCUGGAUUAAAUCUCAAUGGGGGAUAAAUUGUGUUCGGUGGCCGGCAGCCA -------------------------.....(((.(((...(((((((((((........)))))))..-(((((((...(((((.....))))).......)))))))))))))).))). ( -32.10, z-score = -0.82, R) >droYak2.chr2R 12361138 93 - 21139217 -------------------------AGCUGGGCAGCCAAAGGCUGCUGUCACUCGUAUUUGAUGGCAA-GUCGGAUUAAAUCUCAAUGGGG-AUAAAUUGUGUUUGGUGGCCGGCAGCCA -------------------------.(((((.((.((((((((((((((((........))))))).)-))).......(((((....)))-))........))))))).)))))..... ( -33.20, z-score = -1.57, R) >droEre2.scaffold_4845 4965800 93 - 22589142 -------------------------AGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA-GUCGGAUUAAAUCUCAAUGGGG-AUAAAUUUUGUUUGGUGACCAGCAGCCA -------------------------.(((((...(((((((((((((((((........))))))).)-))).......(((((....)))-))........))))))..)))))..... ( -33.90, z-score = -2.68, R) >droAna3.scaffold_13266 5462706 94 + 19884421 -------------------------AGCCAUUUGGCCAAAGGUCGCUGUCAGUCGAUGCUGAUUGCCA-GUCGGAUUAAAUCUCAGGGGAAGAUAAAUUUUAUUUAGUGGCUGGCAGCCA -------------------------.(((....)))....(((....((((((....))))))(((((-((((.......(((....)))(((((.....)))))..)))))))))))). ( -27.10, z-score = -0.46, R) >droWil1.scaffold_180745 661491 118 + 2843958 AGCUGCCAAAUGGGGGUAAAGUUUGAAAUAAAUUUCAUUUCUCUAAUGCCACAAAAUGCCACUGACUGCAUUGCAUCUAAGCCAAUGGAGAGACAAAAGCUAAUCUGGGGAUUAUACA-- .(((.((....)).)))..(((((((((....)))).((((((((.((......(((((........)))))((......)))).))))))))...)))))((((....)))).....-- ( -21.90, z-score = 1.22, R) >consensus _________________________AGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAA_GUUGGAUUAAAUCUCAAUGGGGGAUAAAUUGUGUUUGGUGGCCGGCAGCCA ..........................(((....)))....(((((((((((((...(((((((......)))))))...(((((.....)))))...........)))))).))))))). (-16.66 = -17.30 + 0.64)

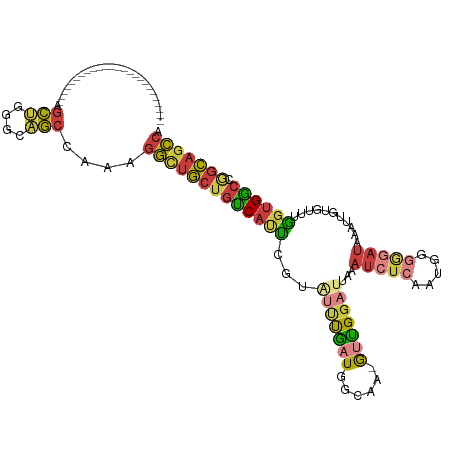
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:29 2011