| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,150,498 – 8,150,671 |
| Length | 173 |
| Max. P | 0.988371 |

| Location | 8,150,498 – 8,150,606 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.01 |
| Shannon entropy | 0.50320 |
| G+C content | 0.50389 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
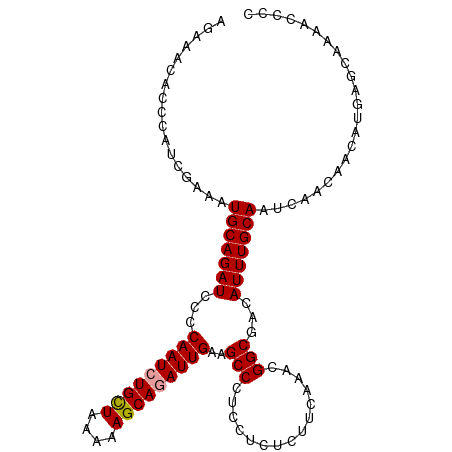
>dm3.chr2R 8150498 108 + 21146708 -UAGCCAGCCCCCCACUCGAUUAACGACCUCGCCCGAUUGACUGCCAUUAAUGGAUAUCAUCGGUGGGUGGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCAUUU -......((.(((((((((.....((....)).(((((.((...(((....)))...)))))))))))))))).((((((((......)))..)))))....))..... ( -30.50, z-score = -0.32, R) >droEre2.scaffold_4845 4946090 103 - 22589142 -----UAGCCCCCUACUCGAUUAACGACCCCGCCCGAUUGACUGCCAUUAAUGGAUAUCAUCGGUGGGUGGGGCUG-GCUGAUGUUUUUGAUUGCAAAUUUCGCCUUUU -----.((((((....(((.....))).((((((.(((.((...(((....)))...))))))))))).))))))(-((.((....((((....))))..))))).... ( -33.90, z-score = -2.21, R) >droYak2.chr2R 12339858 107 - 21139217 -UAGCUAGUCCCCUACUCGAUUAACGACCCCGCCCGAUUGACUGCCAUUAAUGGAUAUCAUCGGUGGGUGGGGCUG-GCUGAUGUUUUUGAUUGCAAAUGUCGCCAUUU -.((((((((((....(((.....))).((((((.(((.((...(((....)))...))))))))))).)))))))-)))(((((((........)))))))....... ( -37.20, z-score = -2.46, R) >droSec1.super_1 5699146 109 + 14215200 AUAGCCAGCCCCCUACUCGAUUAACGACCCCGCCCGAUUGACUGCCAUUAAUGGAUAUUAUCGGUGGGUGGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCGUUU .......((..(.....((((((((((((((((((.(((((...(((....)))......)))))))))))))))..((....)).)))))))).....)..))..... ( -31.00, z-score = -0.58, R) >droSim1.chr2R 6617085 97 + 19596830 ------------CUACUCGAUUAACGACCCCGCCCGAUUGACCGCCAUUAAUGGAUAUCAUCGGUGGGUGGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCGUUU ------------.....((((....(((((((((((..(((...(((....)))...)))....)))))))))))(((((((......)))..))))..))))...... ( -31.30, z-score = -1.47, R) >dp4.chr3 16912892 86 - 19779522 ---------UAUCCAUUCC-CCAACCCUCUCGCGCCACGGAUUGCCAUUAAUGGAUCUGCUGGGUGGGUGGGGU----CUGUUGUUGUUUGCUACCAUUU--------- ---------....((..((-(((.(((.((((.((...((((..(((....))))))))))))).)))))))).----.))...................--------- ( -24.90, z-score = 0.10, R) >droPer1.super_2 5979533 86 - 9036312 ---------UAUCCAUACC-CCAACCCUCUCGCGCCACGGAUUGCCAUUAAUGGAUCUGCUGGGUGGGUGGGGU----CUGUUGUUGUUUGCUACCAUUU--------- ---------....((.(((-(((.(((.((((.((...((((..(((....))))))))))))).)))))))))----.))...................--------- ( -26.20, z-score = -0.23, R) >consensus ______AG_CCCCUACUCGAUUAACGACCCCGCCCGAUUGACUGCCAUUAAUGGAUAUCAUCGGUGGGUGGGGUUU_GCUGAUGUUGUUGAUUGCAAAUGUCGCC_UUU .................((((((((((((((((.(((.(((...(((....)))...)))))))))))...((.....))...)))))))))))............... (-17.71 = -18.24 + 0.54)


| Location | 8,150,566 – 8,150,671 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Shannon entropy | 0.04373 |
| G+C content | 0.45079 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8150566 105 + 21146708 GGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCAUUUGAAGACAGGAGGGCUUCAAUUUGCUUUUCAACAGAUUGGGGAUCUGCAUUUCGAUGGGUGUUUCU (..(.((((((((......)))..)))))...((((..((((((...(((...((((((((((........)))))))))).)))...))))))..)))))..). ( -26.30, z-score = 0.21, R) >droSec1.super_1 5699215 105 + 14215200 GGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCGUUUGAAGAGAGGAGGGCUUCAAUCUGCUUUUUAGCAGAUUGGGGAUCUGCAUUUCGAUGGGUGUUUCU (..(.((((((((......)))..)))))...((((..((((((.(.(((...((((((((((((....)))))))))))).))).).))))))..)))))..). ( -34.30, z-score = -2.05, R) >droSim1.chr2R 6617142 105 + 19596830 GGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCGUUUGAAGAGAGGAGGGCUUCAAUCUGCUUUUUAGCAGAUUGGGGAUCUGCAUUUCGAUGGGUGUUUCU (..(.((((((((......)))..)))))...((((..((((((.(.(((...((((((((((((....)))))))))))).))).).))))))..)))))..). ( -34.30, z-score = -2.05, R) >consensus GGGGUUUUGCUCAUGUUGUUGAUUGCAAAUGUCGCCGUUUGAAGAGAGGAGGGCUUCAAUCUGCUUUUUAGCAGAUUGGGGAUCUGCAUUUCGAUGGGUGUUUCU (..(.((((((((......)))..)))))...((((..((((((...(((...((((((((((((....)))))))))))).)))...))))))..)))))..). (-31.12 = -31.23 + 0.11)

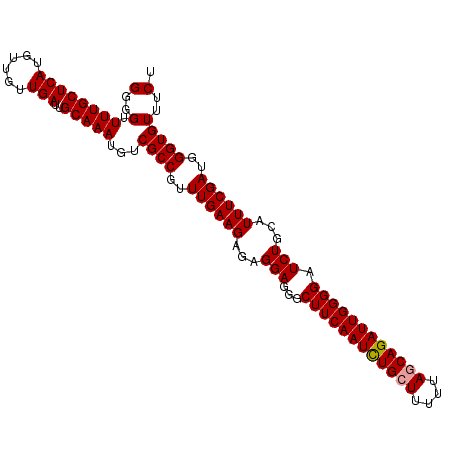
| Location | 8,150,566 – 8,150,671 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Shannon entropy | 0.04373 |
| G+C content | 0.45079 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8150566 105 - 21146708 AGAAACACCCAUCGAAAUGCAGAUCCCCAAUCUGUUGAAAAGCAAAUUGAAGCCCUCCUGUCUUCAAAUGGCGACAUUUGCAAUCAACAACAUGAGCAAAACCCC .................(((((((.....))))(((((...((((((.(..(((....((....))...)))..)))))))..))))).......)))....... ( -18.60, z-score = -1.41, R) >droSec1.super_1 5699215 105 - 14215200 AGAAACACCCAUCGAAAUGCAGAUCCCCAAUCUGCUAAAAAGCAGAUUGAAGCCCUCCUCUCUUCAAACGGCGACAUUUGCAAUCAACAACAUGAGCAAAACCCC .................(((((((...(((((((((....)))))))))..(((...............)))...)))))))....................... ( -21.56, z-score = -3.14, R) >droSim1.chr2R 6617142 105 - 19596830 AGAAACACCCAUCGAAAUGCAGAUCCCCAAUCUGCUAAAAAGCAGAUUGAAGCCCUCCUCUCUUCAAACGGCGACAUUUGCAAUCAACAACAUGAGCAAAACCCC .................(((((((...(((((((((....)))))))))..(((...............)))...)))))))....................... ( -21.56, z-score = -3.14, R) >consensus AGAAACACCCAUCGAAAUGCAGAUCCCCAAUCUGCUAAAAAGCAGAUUGAAGCCCUCCUCUCUUCAAACGGCGACAUUUGCAAUCAACAACAUGAGCAAAACCCC .................(((((((...(((((((((....)))))))))..(((...............)))...)))))))....................... (-19.91 = -20.03 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:25 2011