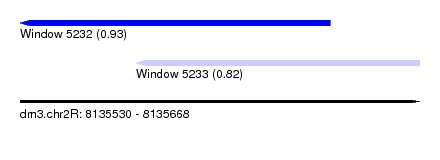
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,135,530 – 8,135,668 |
| Length | 138 |
| Max. P | 0.926419 |

| Location | 8,135,530 – 8,135,637 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Shannon entropy | 0.29113 |
| G+C content | 0.34176 |
| Mean single sequence MFE | -12.64 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
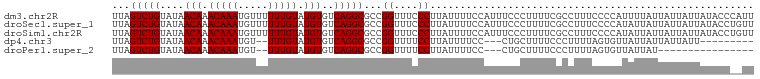
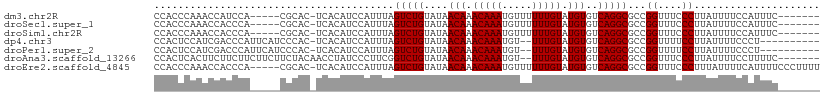
>dm3.chr2R 8135530 107 - 21146708 UUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUCCCUUUUCGCCUUUCCCCAUUUUAUUAUUAUUAUACCCAUU ...(((((....(((.(((((.....))))).)))..)))))...((....))...................................................... ( -12.60, z-score = -1.61, R) >droSec1.super_1 5684275 107 - 14215200 UUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUCCCUUUUCGCCUUUCCCCAUAUUAUUAUUAUUAUACCUGUU ...(((((....(((.(((((.....))))).)))..)))))...((....))...................................................... ( -12.60, z-score = -1.16, R) >droSim1.chr2R 6602050 107 - 19596830 UUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUCCCUUUUCGCCUUUCCCCAUAUUAUUAUUAUUAUACCUGUU ...(((((....(((.(((((.....))))).)))..)))))...((....))...................................................... ( -12.60, z-score = -1.16, R) >dp4.chr3 16897619 93 + 19779522 UUAGUCUGUAUAACAAACAAAUGU--UUUGUAUGUGUCAGGCGCCGGUUUUCCUUAUUUUCC---CUGCUUUUCCCUUUUAGUGUUAUUAUUAUUAUU--------- ...(((((....(((.((((....--.)))).)))..)))))((.((..............)---).))..........(((((....))))).....--------- ( -12.74, z-score = -0.96, R) >droPer1.super_2 5963073 86 + 9036312 UUAGUCUGUAUAACAAACAAAUGU--UUUGUAUGUGUCAGGCGCCGGUUUUCCUUAUUUUCC---CUGCUUUUCCCUUUUAGUGUUAUUAU---------------- ...(((((....(((.((((....--.)))).)))..)))))((.((..............)---).))......................---------------- ( -12.64, z-score = -1.13, R) >consensus UUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUCCCUUUUCGCCUUUCCCCAUAUUAUUAUUAUUAUACC__UU ...(((((....(((.(((((.....))))).)))..)))))...((....))...................................................... (-12.36 = -12.36 + 0.00)
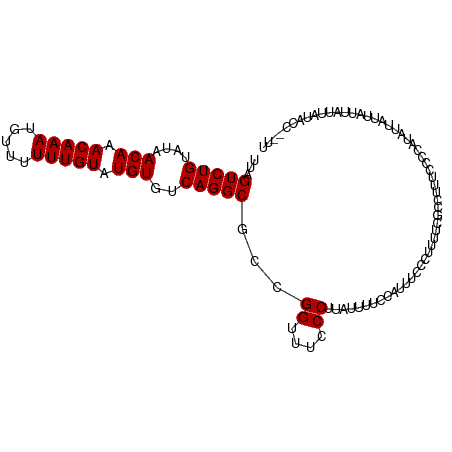
| Location | 8,135,570 – 8,135,668 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Shannon entropy | 0.29882 |
| G+C content | 0.42199 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
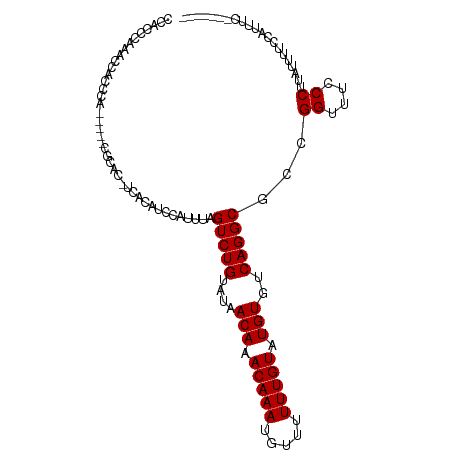
>dm3.chr2R 8135570 98 - 21146708 CCACCCAAACCAUCCA-----CGCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUC------- ......(((((.....-----.(...-...).........(((((....(((.(((((.....))))).)))..)))))...))))).................------- ( -14.70, z-score = -1.54, R) >droSec1.super_1 5684315 98 - 14215200 CCACCCAAACCACCCA-----CGCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUC------- ......(((((.....-----.(...-...).........(((((....(((.(((((.....))))).)))..)))))...))))).................------- ( -14.70, z-score = -1.38, R) >droSim1.chr2R 6602090 98 - 19596830 CCACCCAAACCACCCA-----CGCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUC------- ......(((((.....-----.(...-...).........(((((....(((.(((((.....))))).)))..)))))...))))).................------- ( -14.70, z-score = -1.38, R) >dp4.chr3 16897650 98 + 19779522 CCACUCCAUCGACCCAUUCAUCCCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGU--UUUGUAUGUGUCAGGCGCCGGUUUUCCUUAUUUUCCCU---------- ..........................-.............(((((....(((.((((....--.)))).)))..)))))...((....))...........---------- ( -11.40, z-score = -0.39, R) >droPer1.super_2 5963097 98 + 9036312 CCACUCCAUCGACCCAUUCAUCCCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGU--UUUGUAUGUGUCAGGCGCCGGUUUUCCUUAUUUUCCCU---------- ..........................-.............(((((....(((.((((....--.)))).)))..)))))...((....))...........---------- ( -11.40, z-score = -0.39, R) >droAna3.scaffold_13266 5430910 102 - 19884421 CCACUCACUUCUUCUUCUUCUUCUACAACCUAUCCCUUCGGUCUGUAUAACAAACAAAUGU--UUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCUUUUC------- .....................................((((((((....(((.((((....--.)))).)))..)))..)))))....................------- ( -11.50, z-score = -0.99, R) >droEre2.scaffold_4845 4930329 105 + 22589142 CCACCCAAACCACCCA-----CGCAC-UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUUAUUUUCAUUUUCCCUUUU ......(((((.....-----.(...-...).........(((((....(((.(((((.....))))).)))..)))))...)))))........................ ( -14.70, z-score = -1.54, R) >consensus CCACCCAAACCACCCA_____CGCAC_UCACAUCCAUUUAGUCUGUAUAACAAACAAAUGUUUUUUGUAUGUGUCAGGCGCCGGUUUCCCUUAUUUUCCAUUUC_______ ........................................(((((....(((.(((((.....))))).)))..)))))...((....))..................... (-12.24 = -12.24 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:21 2011