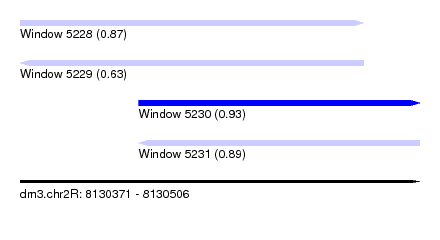
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,130,371 – 8,130,506 |
| Length | 135 |
| Max. P | 0.934678 |

| Location | 8,130,371 – 8,130,487 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
| Shannon entropy | 0.46383 |
| G+C content | 0.45059 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.68 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8130371 116 + 21146708 CGUCGGUUCAGGAACAGAUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAGCUGAAUGCAAAAAGAUACAAAAGUAUCUUAGAAAUACAAGACCAUUU--- .(((.((((((...(.(((((.(((((((((((.((.......))....))))))))..)))))))).))))))).....(((((((....)))))))..........))).....--- ( -31.00, z-score = -2.05, R) >droAna3.scaffold_13266 5426306 100 + 19884421 CAUCGAUUCAGGAACAAUUGUCAGUUGGAACGAACCAAUUGCAAUGCAAGUGGAUCAAGUGAGUCUCAAA-------AAUAGUCUAUAAGGGU-UAUGAGA----CCAAGUU------- ....((((((......((((.(((((((......))))))))))).....))))))...((.((((((..-------..((..((....))..-)))))))----)))....------- ( -25.90, z-score = -2.41, R) >droEre2.scaffold_4845 4925493 110 - 22589142 CAACGGUUCAAGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUAGGCUGAAUCCAAUGAGAUACAAAAGUAUCUCAGAUAUGCCAGA--------- .....(((....)))..((((.(((((((((((.((.......))....))))))))..))))))).(((...(((...((((((((....)))))))))))..)))...--------- ( -32.00, z-score = -1.61, R) >droYak2.chr2R 12319763 119 - 21139217 CAUCGGUUCAAGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUGGCUGAAUCCCAUUAGAUACAAAAGUAUCUUAGAAAUGCCAGAAUACUUGUC .............(((((((.((((((((((((.((.......))....))))))))..))))..(((((....((.....((((((....))))))..))...)))))..))))))). ( -32.00, z-score = -1.13, R) >droSec1.super_1 5679155 104 + 14215200 CGUCGGUUUAGGAACAGGUGCCUCUUGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAGC-------AAUAAGAUACAAAAGUAUCUUAGAAAUACAAGAC-------- .((((((............))).((((((((((.((.......))....))))))))))...(((((...-------..((((((((....))))))))..)))))..)))-------- ( -29.00, z-score = -1.53, R) >droSim1.chr2R 6596494 109 + 19596830 CGUCGGUUUAGGAACAGGUGCCUCUUGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAGC-------AAUAAGAUACAAAAGUAUCUUAGAAAUACAAGACCAUUU--- ....(((((.(.....((..((.((((((((((.((.......))....)))))))))).).)..))...-------..((((((((....))))))))......).)))))....--- ( -32.10, z-score = -2.11, R) >consensus CAUCGGUUCAGGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAGC_______AAUAAGAUACAAAAGUAUCUUAGAAAUACAAGAC________ ...(((((......(((..((((........))))...)))......)))))...........................((((((((....)))))))).................... (-15.98 = -15.68 + -0.30)

| Location | 8,130,371 – 8,130,487 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Shannon entropy | 0.46383 |
| G+C content | 0.45059 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8130371 116 - 21146708 ---AAAUGGUCUUGUAUUUCUAAGAUACUUUUGUAUCUUUUUGCAUUCAGCUAAUACUCACUUGAAACGGUUGUCCGGCAGUUGGCCCGCUCCAUGAGGCAUCUGUUCCUGAACCGACG ---.....(((.((((.....(((((((....)))))))..))))(((((.................(((....)))((((...(((((.....)).)))..))))..)))))..))). ( -27.40, z-score = -1.02, R) >droAna3.scaffold_13266 5426306 100 - 19884421 -------AACUUGG----UCUCAUA-ACCCUUAUAGACUAUU-------UUUGAGACUCACUUGAUCCACUUGCAUUGCAAUUGGUUCGUUCCAACUGACAAUUGUUCCUGAAUCGAUG -------.....((----(((((..-................-------..)))))))...(((((.((...((((((((.((((......)))).)).))).)))...)).))))).. ( -18.95, z-score = -1.43, R) >droEre2.scaffold_4845 4925493 110 + 22589142 ---------UCUGGCAUAUCUGAGAUACUUUUGUAUCUCAUUGGAUUCAGCCUAUACUCACUUGAAACGGUUGUCCGGCAGUUGGCCCGCUCCAUGAGGCACCUGUUCUUGAACCGUUG ---------...(((..(((((((((((....)))))))...))))...))).............(((((((....(((((...(((((.....)).)))..)))))....))))))). ( -32.20, z-score = -1.78, R) >droYak2.chr2R 12319763 119 + 21139217 GACAAGUAUUCUGGCAUUUCUAAGAUACUUUUGUAUCUAAUGGGAUUCAGCCAAUACUCACUUGAAACGGUUGUCCGGCAGUUGGCCCGCUCCAUGAGGCACCUGUUCUUGAACCGAUG ..(((((....((((..(((((((((((....))))))..)))))....))))......)))))...(((((....(((((...(((((.....)).)))..)))))....)))))... ( -32.20, z-score = -1.18, R) >droSec1.super_1 5679155 104 - 14215200 --------GUCUUGUAUUUCUAAGAUACUUUUGUAUCUUAUU-------GCUAACACUCACUUGAAACGGUCGUCCGGCAGUUGGCCCGCUCCAAGAGGCACCUGUUCCUAAACCGACG --------(((..(((....((((((((....)))))))).)-------)).................(((.....(((((...((((.......).)))..))))).....)))))). ( -23.70, z-score = -1.09, R) >droSim1.chr2R 6596494 109 - 19596830 ---AAAUGGUCUUGUAUUUCUAAGAUACUUUUGUAUCUUAUU-------GCUAACACUCACUUGAAACGGUCGUCCGGCAGUUGGCCCGCUCCAAGAGGCACCUGUUCCUAAACCGACG ---...((((((((......((((((((....))))))))..-------((((((..((....))..(((....)))...))))))......))))(((........)))..))))... ( -24.30, z-score = -0.63, R) >consensus ________GUCUGGUAUUUCUAAGAUACUUUUGUAUCUUAUU_______GCUAAUACUCACUUGAAACGGUUGUCCGGCAGUUGGCCCGCUCCAAGAGGCACCUGUUCCUGAACCGACG ....................((((((((....))))))))...........................(((((....(((((...(((..........)))..)))))....)))))... (-15.95 = -16.82 + 0.86)


| Location | 8,130,411 – 8,130,506 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 68.33 |
| Shannon entropy | 0.50223 |
| G+C content | 0.36831 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -11.06 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
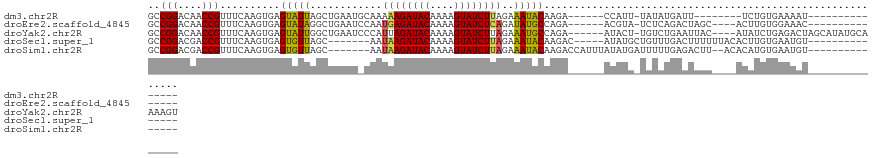
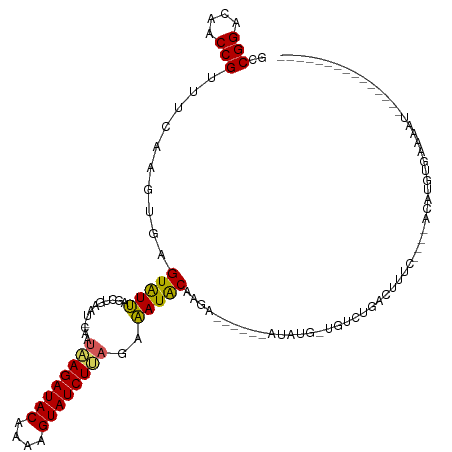
>dm3.chr2R 8130411 95 + 21146708 GCCGGACAACCGUUUCAAGUGAGUAUUAGCUGAAUGCAAAAAGAUACAAAAGUAUCUUAGAAAUACAAGA------CCAUU-UAUAUGAUU--------UCUGUGAAAAU--------------- ..(((....)))(((((...(((((((.((.....))...(((((((....)))))))...)))))....------.(((.-...)))...--------))..)))))..--------------- ( -17.20, z-score = -1.72, R) >droEre2.scaffold_4845 4925533 99 - 22589142 GCCGGACAACCGUUUCAAGUGAGUAUAGGCUGAAUCCAAUGAGAUACAAAAGUAUCUCAGAUAUGCCAGA------ACGUA-UCUCAGACUAGC----ACUUGUGGAAAC--------------- .((((....))....((((((....(((.((((......((((((((....))))))))((((((.....------.))))-)))))).))).)----))))).))....--------------- ( -31.00, z-score = -3.76, R) >droYak2.chr2R 12319803 114 - 21139217 GCCGGACAACCGUUUCAAGUGAGUAUUGGCUGAAUCCCAUUAGAUACAAAAGUAUCUUAGAAAUGCCAGA------AUACU-UGUCUGAAUUAC----AUAUCUGAGACUAGCAUAUGCAAAAGU ..(((....))).((((.(..(((((((((....((.....((((((....))))))..))...))))..------)))))-..).))))...(----(((((((....))).)))))....... ( -24.40, z-score = -1.38, R) >droSec1.super_1 5679195 98 + 14215200 GCCGGACGACCGUUUCAAGUGAGUGUUAGC-------AAUAAGAUACAAAAGUAUCUUAGAAAUACAAGAC-----AUAUGCUGUUUGACUUUUUUACACUUGUGAAUGU--------------- ..........(((((((((((.(((((...-------..((((((((....))))))))..))))).((((-----(.....)))))..........)))))).))))).--------------- ( -22.00, z-score = -1.56, R) >droSim1.chr2R 6596534 101 + 19596830 GCCGGACGACCGUUUCAAGUGAGUGUUAGC-------AAUAAGAUACAAAAGUAUCUUAGAAAUACAAGACCAUUUAUAUGAUUUUUGAGACUU--ACACAUGUGAAUGU--------------- ..(((....)))......((((((.((((.-------..((((((((....))))))))............(((....)))....)))).))))--))((((....))))--------------- ( -20.30, z-score = -1.71, R) >consensus GCCGGACAACCGUUUCAAGUGAGUAUUAGCUGAAU_CAAUAAGAUACAAAAGUAUCUUAGAAAUACAAGA______AUAUG_UGUCUGACUUUC____ACAUGUGAAAAU_______________ ..(((....)))..........(((((............((((((((....))))))))..)))))........................................................... (-11.06 = -10.42 + -0.64)
| Location | 8,130,411 – 8,130,506 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 68.33 |
| Shannon entropy | 0.50223 |
| G+C content | 0.36831 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8130411 95 - 21146708 ---------------AUUUUCACAGA--------AAUCAUAUA-AAUGG------UCUUGUAUUUCUAAGAUACUUUUGUAUCUUUUUGCAUUCAGCUAAUACUCACUUGAAACGGUUGUCCGGC ---------------..(((((.((.--------.(((((...-.))))------)..((((.....(((((((....)))))))..))))...............)))))))(((....))).. ( -15.60, z-score = -0.89, R) >droEre2.scaffold_4845 4925533 99 + 22589142 ---------------GUUUCCACAAGU----GCUAGUCUGAGA-UACGU------UCUGGCAUAUCUGAGAUACUUUUGUAUCUCAUUGGAUUCAGCCUAUACUCACUUGAAACGGUUGUCCGGC ---------------.......(((((----(.(((.((((((-((.((------....)).))))((((((((....))))))))......)))).)))....))))))...(((....))).. ( -28.70, z-score = -2.32, R) >droYak2.chr2R 12319803 114 + 21139217 ACUUUUGCAUAUGCUAGUCUCAGAUAU----GUAAUUCAGACA-AGUAU------UCUGGCAUUUCUAAGAUACUUUUGUAUCUAAUGGGAUUCAGCCAAUACUCACUUGAAACGGUUGUCCGGC ....((((((((.((......))))))----))))(((((...-(((((------..((((..(((((((((((....))))))..)))))....)))))))))...))))).(((....))).. ( -28.80, z-score = -2.14, R) >droSec1.super_1 5679195 98 - 14215200 ---------------ACAUUCACAAGUGUAAAAAAGUCAAACAGCAUAU-----GUCUUGUAUUUCUAAGAUACUUUUGUAUCUUAUU-------GCUAACACUCACUUGAAACGGUCGUCCGGC ---------------.......((((((......((.(((...(((...-----....))).....((((((((....))))))))))-------)))......))))))...(((....))).. ( -20.60, z-score = -2.08, R) >droSim1.chr2R 6596534 101 - 19596830 ---------------ACAUUCACAUGUGU--AAGUCUCAAAAAUCAUAUAAAUGGUCUUGUAUUUCUAAGAUACUUUUGUAUCUUAUU-------GCUAACACUCACUUGAAACGGUCGUCCGGC ---------------...((((...((((--.(((.......(((((....)))))..........((((((((....))))))))..-------))).)))).....)))).(((....))).. ( -19.50, z-score = -2.03, R) >consensus _______________ACUUUCACAAGU____AAAAGUCAAACA_AAUAU______UCUUGUAUUUCUAAGAUACUUUUGUAUCUUAUUG_AUUCAGCUAAUACUCACUUGAAACGGUUGUCCGGC ..................................................................((((((((....))))))))...........................(((....))).. (-10.76 = -10.92 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:19 2011