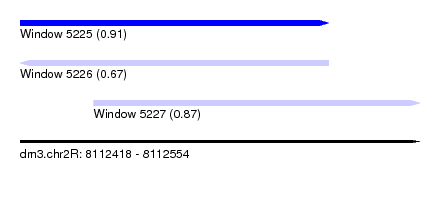
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,112,418 – 8,112,554 |
| Length | 136 |
| Max. P | 0.907758 |

| Location | 8,112,418 – 8,112,523 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Shannon entropy | 0.43338 |
| G+C content | 0.58943 |
| Mean single sequence MFE | -45.35 |
| Consensus MFE | -25.13 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
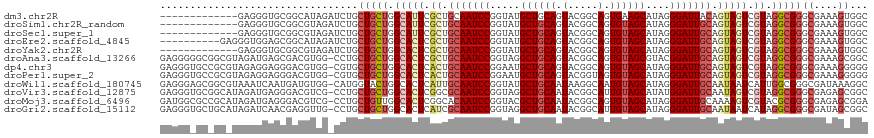
>dm3.chr2R 8112418 105 + 21146708 -------------GAGGGUGCGGCAUAGAUCUGCUGCUGGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGAAGCAUAGGGAUUACAGUAGUCGUAGGCGGGCGAAAGUGGC -------------......((((((......))))))..((...(((((((((((..((((((....(((....))).)))))).)))))...)))).))...))..((....))... ( -32.00, z-score = 1.04, R) >droSim1.chr2R_random 2216633 105 + 2996586 -------------GAGGGUGCGGCGUAGAUCUGCUGCUGGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGC -------------..((((((((((((....))).))).))))))((((((((((..(((((((((.(.....).))))))))).))))))))))..(((....)))((....))... ( -43.40, z-score = -1.93, R) >droSec1.super_1 5661076 105 + 14215200 -------------GAGGGUGCGGCGUAGAUCUGCUGCUGGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGC -------------..((((((((((((....))).))).))))))((((((((((..(((((((((.(.....).))))))))).))))))))))..(((....)))((....))... ( -43.40, z-score = -1.93, R) >droEre2.scaffold_4845 4905292 108 - 22589142 ----------GAGGGUGGAGCGGCAUAGAUCUGCUGCUGGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGC ----------..(((((.(((((((......)))))))..)))))((((((((((..(((((((((.(.....).))))))))).))))))))))..(((....)))((....))... ( -48.00, z-score = -3.17, R) >droYak2.chr2R 12301264 105 - 21139217 -------------GAGGGUGCGGCGUAGAUCUGCUGCUGGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGC -------------..((((((((((((....))).))).))))))((((((((((..(((((((((.(.....).))))))))).))))))))))..(((....)))((....))... ( -45.80, z-score = -2.30, R) >droAna3.scaffold_13266 5404214 117 + 19884421 GAGGGGGCGGCGUAGAUGAGCGACGUGG-CCUGCUGCUGGCACUCGCUGCAAUCCGGUAGGCUGCAGUACGGCAGUGUAGCGUACGGAUUGCAGUAGUCGUAGGCGGGCGAAAGCGGC ....((((.((((.(.....).)))).)-))).(((((.(((((.(((((((((((.((.((((((.(.....).)))))).)))))))))))))))).)).)))))((....))... ( -57.10, z-score = -2.97, R) >dp4.chr3 16873904 117 - 19779522 GAGGGUGCCGCGUAGAGGAGGGACGUGG-CGUGCUGCUGGCACUCACUGCAAUCCGGAAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGGGGG ......(((((((.........))))))-)...(((((.(((((.((((((((((...((((((((.(.....).))))))))..))))))))))))).)).))))).(....).... ( -49.60, z-score = -2.87, R) >droPer1.super_2 5939857 117 - 9036312 GAGGGUGCCGCGUAGAGGAGGGACGUGG-CGUGCUGCUGGCACUCACUGCAAUCCGGAAUGCUGCAGUACGGUAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGGGGG ......(((((((.........))))))-)...(((((.(((((.((((((((((...((((((((.((...)).))))))))..))))))))))))).)).))))).(....).... ( -50.00, z-score = -3.34, R) >droWil1.scaffold_180745 564556 117 + 2843958 GAGGGAGCGGCGUAAAUCAAUGAUGUGG-CAUGGUACUGGCACUCAUUGCAAUCCGGUAUGCUGCAAUAAGGCAAUGUAGCAUAGGGAUUGCAAUAAUCAUUGGCGGGCGAUAAAGGC ..........(((...(((((((((((.-((......)).)))..((((((((((..(((((((((.........))))))))).)))))))))).))))))))...)))........ ( -39.90, z-score = -2.83, R) >droVir3.scaffold_12875 13864680 117 - 20611582 GAGGGUGCGGCAUAGAUGAGGGACGUCG-CCUGCUGCUGGCACUCGGCGCAAUCCGGUAGGCUGCAAUACGGCAUUGUAGCAUAUGGAUUGCAAUAGUCGUAGGCGGGCGAGAGCGGC ......((.((...((((.....))))(-((((((((((.....))))((((((((....((((((((.....))))))))...))))))))..........)))))))....)).)) ( -49.10, z-score = -2.09, R) >droMoj3.scaffold_6496 13474035 117 + 26866924 GAUGGCGCCGCAUAGAUGAGGGACGUCG-CCUGCUGUUGGCACUCGGCACAAUCCGGUACGCUGCAAUACGGCAGUGUAGCAUAGGGAUUGCAAAAGUCGUACGCGGGCGAGAGCGGA .......((((...((((.....))))(-(((((.((((((.....(((..(((((.((((((((......)))))))).)....)))))))....)))).))))))))....)))). ( -44.70, z-score = -0.74, R) >droGri2.scaffold_15112 3551444 117 - 5172618 GAGGGUGCUGCAUAGAUCAACGAGGUUG-CCUGCUGCUGGCACUCAUCGCAAUCCGGUAGGCUGCAAUACGGCAUUGUAGCAUAGGGAUUGCAAUAAUCAUAGGCGGGCGAUAGCGGC ......(((((...((((.....))))(-(((((((....).......(((((((.....((((((((.....))))))))....)))))))..........)))))))....))))) ( -41.20, z-score = -0.77, R) >consensus GAGGG_GC_GC_UAGAGGAGCGACGUAG_CCUGCUGCUGGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGC .................................(((((.(((((.((.(((((((.....((((((.(.....).))))))....))))))).))))).)).)))))((....))... (-25.13 = -25.77 + 0.64)

| Location | 8,112,418 – 8,112,523 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Shannon entropy | 0.43338 |
| G+C content | 0.58943 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -17.05 |
| Energy contribution | -16.30 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8112418 105 - 21146708 GCCACUUUCGCCCGCCUACGACUACUGUAAUCCCUAUGCUUCACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAAUGCCAGCAGCAGAUCUAUGCCGCACCCUC------------- ((....(((((.((....)).....((((((((.((((((.((.........)).))))))..))))))))))))).))..((.(((......))).))......------------- ( -27.00, z-score = -1.38, R) >droSim1.chr2R_random 2216633 105 - 2996586 GCCACUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAAUGCCAGCAGCAGAUCUACGCCGCACCCUC------------- ((....(((((.((....)).....((((((((.((((((.((.........)).))))))..))))))))))))).))..((.((........)).))......------------- ( -27.10, z-score = -1.46, R) >droSec1.super_1 5661076 105 - 14215200 GCCACUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAAUGCCAGCAGCAGAUCUACGCCGCACCCUC------------- ((....(((((.((....)).....((((((((.((((((.((.........)).))))))..))))))))))))).))..((.((........)).))......------------- ( -27.10, z-score = -1.46, R) >droEre2.scaffold_4845 4905292 108 + 22589142 GCCACUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAGUGCCAGCAGCAGAUCUAUGCCGCUCCACCCUC---------- (.((((((((........)))...(((((((((.((((((.((.........)).))))))..))))))))).))))).)(((.(((......))).)))........---------- ( -30.80, z-score = -2.04, R) >droYak2.chr2R 12301264 105 + 21139217 GCCACUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAGUGCCAGCAGCAGAUCUACGCCGCACCCUC------------- (.((((((((........)))...(((((((((.((((((.((.........)).))))))..))))))))).))))).).((.((........)).))......------------- ( -28.80, z-score = -1.43, R) >droAna3.scaffold_13266 5404214 117 - 19884421 GCCGCUUUCGCCCGCCUACGACUACUGCAAUCCGUACGCUACACUGCCGUACUGCAGCCUACCGGAUUGCAGCGAGUGCCAGCAGCAGG-CCACGUCGCUCAUCUACGCCGCCCCCUC ((.(((..(((.(((..........(((((((((((.(((.((.........)).))).)).)))))))))))).)))..))).)).((-(..(((.(.....).)))..)))..... ( -33.70, z-score = -0.30, R) >dp4.chr3 16873904 117 + 19779522 CCCCCUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUUCCGGAUUGCAGUGAGUGCCAGCAGCACG-CCACGUCCCUCCUCUACGCGGCACCCUC .......(((........))).(((((((((((..(((((.((.........)).)))))...))))))))))).((((.....))))(-((.(((.........))).)))...... ( -35.00, z-score = -2.98, R) >droPer1.super_2 5939857 117 + 9036312 CCCCCUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUACCGUACUGCAGCAUUCCGGAUUGCAGUGAGUGCCAGCAGCACG-CCACGUCCCUCCUCUACGCGGCACCCUC .......(((........))).(((((((((((..(((((.((.........)).)))))...))))))))))).((((.....))))(-((.(((.........))).)))...... ( -35.00, z-score = -3.57, R) >droWil1.scaffold_180745 564556 117 - 2843958 GCCUUUAUCGCCCGCCAAUGAUUAUUGCAAUCCCUAUGCUACAUUGCCUUAUUGCAGCAUACCGGAUUGCAAUGAGUGCCAGUACCAUG-CCACAUCAUUGAUUUACGCCGCUCCCUC ...............((((((((((((((((((.((((((.((.........)).))))))..))))))))))))(((.((......))-.))).))))))................. ( -32.00, z-score = -3.53, R) >droVir3.scaffold_12875 13864680 117 + 20611582 GCCGCUCUCGCCCGCCUACGACUAUUGCAAUCCAUAUGCUACAAUGCCGUAUUGCAGCCUACCGGAUUGCGCCGAGUGCCAGCAGCAGG-CGACGUCCCUCAUCUAUGCCGCACCCUC ((.((..(((((.((((...(((..((((((((.((.(((.(((((...))))).))).))..))))))))...)))...))..)).))-))).(.....)......)).))...... ( -30.40, z-score = 0.04, R) >droMoj3.scaffold_6496 13474035 117 - 26866924 UCCGCUCUCGCCCGCGUACGACUUUUGCAAUCCCUAUGCUACACUGCCGUAUUGCAGCGUACCGGAUUGUGCCGAGUGCCAACAGCAGG-CGACGUCCCUCAUCUAUGCGGCGCCAUC .........(((((((((.((.....(((((((....(.(((.((((......)))).))).))))))))(((...(((.....)))))-)...........))))))))).)).... ( -34.10, z-score = -0.02, R) >droGri2.scaffold_15112 3551444 117 + 5172618 GCCGCUAUCGCCCGCCUAUGAUUAUUGCAAUCCCUAUGCUACAAUGCCGUAUUGCAGCCUACCGGAUUGCGAUGAGUGCCAGCAGCAGG-CAACCUCGUUGAUCUAUGCAGCACCCUC ((.((....))..))......((((((((((((....(((.(((((...))))).))).....))))))))))))((((..(((..(((-((((...)))).))).))).)))).... ( -33.00, z-score = -0.81, R) >consensus GCCACUUUCGCCCGCCUACGACUACUGCAAUCCCUAUGCUACACUGCCGUACUGCAGCAUACCGGAUUGCAGCGAGUGCCAGCAGCAGG_CUACGUCGCUCCUCUA_GC_GC_CCCUC .........((..((...((....(((((((((....(((.((.........)).))).....)))))))))))...))..))................................... (-17.05 = -16.30 + -0.75)

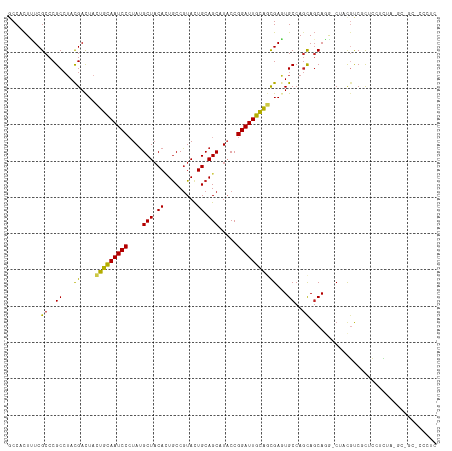
| Location | 8,112,443 – 8,112,554 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Shannon entropy | 0.30127 |
| G+C content | 0.60123 |
| Mean single sequence MFE | -43.43 |
| Consensus MFE | -26.53 |
| Energy contribution | -26.46 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8112443 111 + 21146708 GGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGAAGCAUAGGGAUUACAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGUGCGGGGAAAUCGAUCUGCGGCGGAG--------- ....(((((((((..((.(((((((((..((((((.((((..(....)..))))....)))))).....((....)).))))).)))).))((......))))))))))).--------- ( -36.80, z-score = -0.23, R) >droSim1.chr2R_random 2216658 111 + 2996586 GGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGUGCGGGGAAAUCGAUCUGCGGCGGAG--------- ....(((((((((((((..(((((((((.(.....).))))))))).)))))))))).(((((((((((((....)).(((....))).......))).))))))))))).--------- ( -46.80, z-score = -2.96, R) >droSec1.super_1 5661101 111 + 14215200 GGCAUUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGUGCGGGAAAAUCGAUCUGCGGCGGAG--------- ....(((((((((((((..(((((((((.(.....).))))))))).)))))))))).(((((((((((((....)).(((....))).......))).))))))))))).--------- ( -46.80, z-score = -3.08, R) >droEre2.scaffold_4845 4905320 111 - 22589142 GGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGGGCGGGGAAUUCGAUCUGCGGCGGAG--------- ....(((((((((((((..(((((((((.(.....).))))))))).)))))))))).(((((((((((((....)).((......)).......))).)))))))).)))--------- ( -48.10, z-score = -3.14, R) >droYak2.chr2R 12301289 111 - 21139217 GGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGGGCGGGGAACUCGAUCUGCGGCGGAG--------- ....(((((((((((((..(((((((((.(.....).))))))))).)))))))))).(((((((((.(.(....)).))..((((........))))..))))))).)))--------- ( -49.40, z-score = -3.07, R) >droAna3.scaffold_13266 5404251 111 + 19884421 GGCACUCGCUGCAAUCCGGUAGGCUGCAGUACGGCAGUGUAGCGUACGGAUUGCAGUAGUCGUAGGCGGGCGAAAGCGGCGGUGGGGCGGGGAAAUCGAUCUGCGGAGGCG--------- .((.((((((((((((((.((.((((((.(.....).)))))).)))))))))))))...(((((((((((....)).((......)).......))).))))))))))).--------- ( -51.40, z-score = -3.71, R) >dp4.chr3 16873941 111 - 19779522 GGCACUCACUGCAAUCCGGAAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGGGGGGGCGGGGCGGGAAAGUCGAUUUGCGGUGGUG--------- .((.(((((((((((((...((((((((.(.....).))))))))..))))))))))..((((......))))..)))...))...((.(.(((.....))).).))....--------- ( -37.80, z-score = -1.55, R) >droPer1.super_2 5939894 111 - 9036312 GGCACUCACUGCAAUCCGGAAUGCUGCAGUACGGUAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGGGGGGGCGGGGCGGGAAAGUCGAUUUGCGGUGGUG--------- .((.(((((((((((((...((((((((.((...)).))))))))..))))))))))..((((......))))..)))...))...((.(.(((.....))).).))....--------- ( -38.20, z-score = -2.12, R) >droWil1.scaffold_180745 564593 107 + 2843958 GGCACUCAUUGCAAUCCGGUAUGCUGCAAUAAGGCAAUGUAGCAUAGGGAUUGCAAUAAUCAUUGGCGGGCGAUAAAGGCGGUGGAGCUGGGAAAUCAAUCUGUGGU------------- (((....((((((((((..(((((((((.........))))))))).))))))))))..((((((.(..........).)))))).)))..................------------- ( -32.60, z-score = -1.37, R) >droVir3.scaffold_12875 13864717 111 - 20611582 GGCACUCGGCGCAAUCCGGUAGGCUGCAAUACGGCAUUGUAGCAUAUGGAUUGCAAUAGUCGUAGGCGGGCGAGAGCGGCGGCGGUGCGGGGAAGUCAAUCUGUGGCGUCG--------- .((.((((.(((((((((....((((((((.....))))))))...))))))))....((.....)).).)))).))..(((((.((((((........)))))).)))))--------- ( -47.00, z-score = -2.73, R) >droMoj3.scaffold_6496 13474072 111 + 26866924 GGCACUCGGCACAAUCCGGUACGCUGCAAUACGGCAGUGUAGCAUAGGGAUUGCAAAAGUCGUACGCGGGCGAGAGCGGAGGCGGCGCGGGGAAAUCAAUCUGUGGCAUUG--------- .((.(((.((.(((((((.((((((((......)))))))).)....))))))......((((......))))..)).)))))(.((((((........)))))).)....--------- ( -41.60, z-score = -2.11, R) >droGri2.scaffold_15112 3551481 120 - 5172618 GGCACUCAUCGCAAUCCGGUAGGCUGCAAUACGGCAUUGUAGCAUAGGGAUUGCAAUAAUCAUAGGCGGGCGAUAGCGGCGGCGGUGCAGGAAACUCAAUCUGUGGUGGUGCGGGUGGUG ..(((((...(((((((.....((((((((.....))))))))....)))))))....(((((((((.(.((....)).).))..((.((....))))..))))))).....)))))... ( -44.60, z-score = -2.05, R) >consensus GGCACUCGCUGCAAUCCGGUAUGCUGCAGUACGGCAGUGUAGCAUAGGGAUUGCAGUAGUCGUAGGCGGGCGAAAGUGGCGGCGGGGCGGGGAAAUCGAUCUGCGGCGGAG_________ .((..((((((((((((.....((((((.(.....).))))))....))))))).....((((......)))).......))))).(((((........))))).))............. (-26.53 = -26.46 + -0.07)

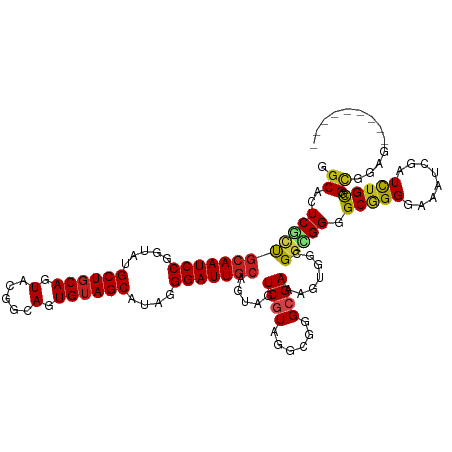
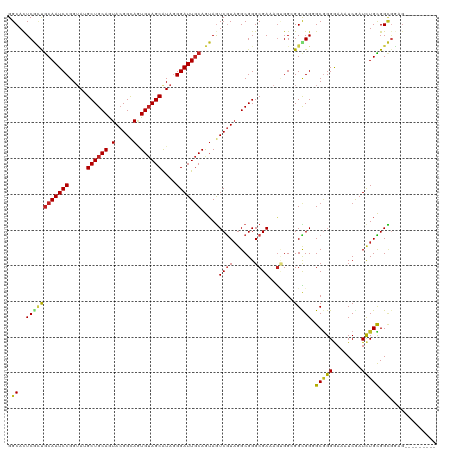
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:16 2011