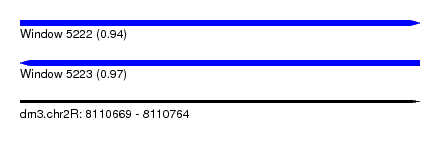
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,110,669 – 8,110,764 |
| Length | 95 |
| Max. P | 0.970165 |

| Location | 8,110,669 – 8,110,764 |
|---|---|
| Length | 95 |
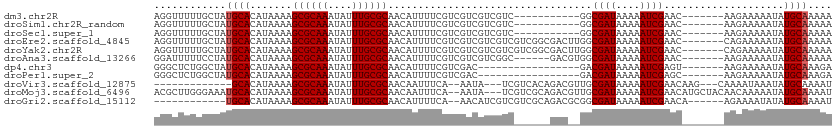
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.44523 |
| G+C content | 0.37890 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.939975 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
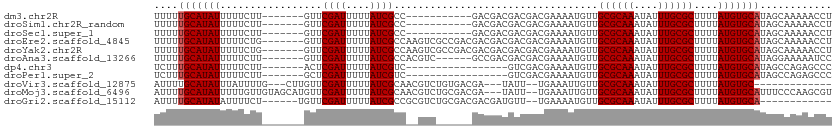
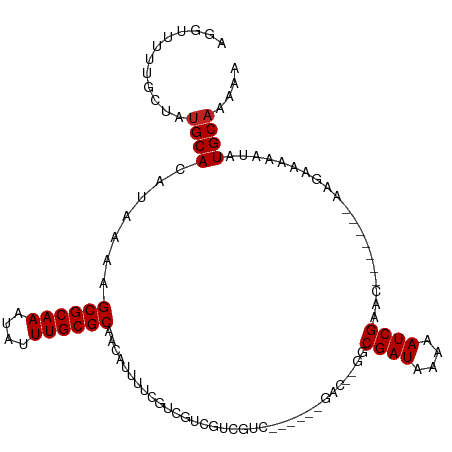
>dm3.chr2R 8110669 95 + 21146708 UUUUUGCAUAUUUUUCUU-------GUUCGAUUUUUAUCGCC-----------GACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU (((((((((((((((.((-------(((((((....))))..-----------......))))).)))))))).((((((....)))))).............)))))))... ( -22.30, z-score = -1.43, R) >droSim1.chr2R_random 2210046 95 + 2996586 UUUUUGCAUAUUUUUCUU-------GUUCGAUUUUUAUCGCC-----------GACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU (((((((((((((((.((-------(((((((....))))..-----------......))))).)))))))).((((((....)))))).............)))))))... ( -22.30, z-score = -1.43, R) >droSec1.super_1 5659430 95 + 14215200 UUUUUGCAUAUUUUUCUU-------GUUCGAUUUUUAUCGCC-----------GACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU (((((((((((((((.((-------(((((((....))))..-----------......))))).)))))))).((((((....)))))).............)))))))... ( -22.30, z-score = -1.43, R) >droEre2.scaffold_4845 4903677 106 - 22589142 UUUUUGCAUAUUUUUCUG-------GUUCGAUUUUUAUCGCCAAGUCGCCGACGACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU (((((((((((((((.((-------(..((((....))))))).((((.((....)).))))...)))))))).((((((....)))))).............)))))))... ( -27.70, z-score = -1.44, R) >droYak2.chr2R 12299685 106 - 21139217 UUUUUGCAUAUUUUUCUG-------GUUCGAUUUUUAUCGCCAAGUCGCCGACGACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU (((((((((((((((.((-------(..((((....))))))).((((.((....)).))))...)))))))).((((((....)))))).............)))))))... ( -27.70, z-score = -1.44, R) >droAna3.scaffold_13266 5402647 100 + 19884421 UUUUUGCAUAUUUUUCUU-------GUUCGAUUUUUAUCGCCACGUC------GCCGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGGAAAAAUCC ....((((((((((((..-------...((((....))))...((((------(....)))))..)))))....((((((....))))))....)))))))..(((....))) ( -25.70, z-score = -1.66, R) >dp4.chr3 16872205 89 - 19779522 UCUUUGCAUAUUUUUCUU-------ACUCGAUUUUUAUCGUC-----------------GUCGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCCAGAGCCC .((.(((((((.......-------.....(((((..(((..-----------------..)))..)))))...((((((....))))))....))))))).))......... ( -20.90, z-score = -1.78, R) >droPer1.super_2 5938211 89 - 9036312 UCUUUGCAUAUUUUUCUU-------GCUCGAUUUUUAUCGUC-----------------GUCGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCCAGAGCCC .((((((.........((-------((.((((.(((.(((((-----------------...)))))))).)))).)))).....(((((......)))))..).)))))... ( -21.60, z-score = -1.49, R) >droVir3.scaffold_12875 13860673 92 - 20611582 AUUUUGCAUAUUUAUUUUG---CUUGUUCGAUUUUUAUCGCAACGUCUGUGACGA---UAUU--UGAAAUUGUUGCGCAAAUAUUUGCGCUUUUAUGUGC------------- .....((((((.......(---(...(((((...(..(((((.....)))))..)---...)--))))...)).((((((....))))))....))))))------------- ( -21.30, z-score = -1.56, R) >droMoj3.scaffold_6496 13470264 108 + 26866924 AUUUUGCAUAUUUUUGUUGUAGCAUGUUCGAUUUUUAUCGCAACGUCUGCGACGA---UAUU--UGAAAUUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUUUCCCAAGCGU ....(((((((..(((((((((.(((((((((....)))).))))))))))))))---....--..........((((((....))))))....)))))))............ ( -32.40, z-score = -3.09, R) >droGri2.scaffold_15112 3548384 93 - 5172618 AUUUUGCAUAUAUUUUCU------UGUUCGAUUUUUAUCGCCGCGUCUGCGACGACGAUGUU--UGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCA------------ ....((((((((......------..(((((....(((((.((((....)).)).))))).)--))))......((((((....))))))...))))))))------------ ( -26.60, z-score = -2.11, R) >consensus UUUUUGCAUAUUUUUCUU_______GUUCGAUUUUUAUCGCC__GUC______GACGACGACGACGAAAAUGUUGCGCAAAUAUUUGCGCUUUUAUGUGCAUAGCAAAAACCU ....(((((((.................((((....))))..................................((((((....))))))....)))))))............ (-15.98 = -16.07 + 0.09)
| Location | 8,110,669 – 8,110,764 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.44523 |
| G+C content | 0.37890 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -14.45 |
| Energy contribution | -14.54 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970165 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
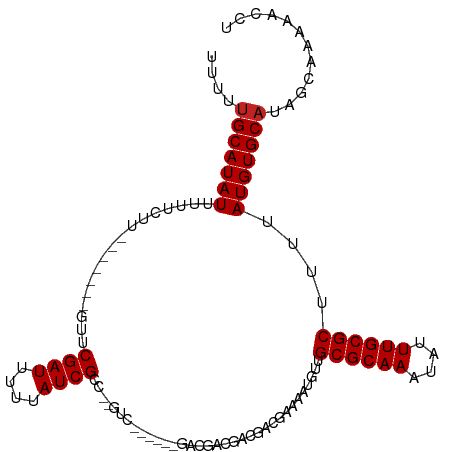
>dm3.chr2R 8110669 95 - 21146708 AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUC-----------GGCGAUAAAAAUCGAAC-------AAGAAAAAUAUGCAAAAA ...((((((((((..........((((((....))))))...(((((..((((((....)-----------)))))..))))).....-------.......))).))))))) ( -25.10, z-score = -2.23, R) >droSim1.chr2R_random 2210046 95 - 2996586 AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUC-----------GGCGAUAAAAAUCGAAC-------AAGAAAAAUAUGCAAAAA ...((((((((((..........((((((....))))))...(((((..((((((....)-----------)))))..))))).....-------.......))).))))))) ( -25.10, z-score = -2.23, R) >droSec1.super_1 5659430 95 - 14215200 AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUC-----------GGCGAUAAAAAUCGAAC-------AAGAAAAAUAUGCAAAAA ...((((((((((..........((((((....))))))...(((((..((((((....)-----------)))))..))))).....-------.......))).))))))) ( -25.10, z-score = -2.23, R) >droEre2.scaffold_4845 4903677 106 + 22589142 AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUCGUCGGCGACUUGGCGAUAAAAAUCGAAC-------CAGAAAAAUAUGCAAAAA ...((((((((((..........((((((....))))))....(((((...(((((((....))))))).(((((((....))))..)-------)))))))))).))))))) ( -31.90, z-score = -2.55, R) >droYak2.chr2R 12299685 106 + 21139217 AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUCGUCGGCGACUUGGCGAUAAAAAUCGAAC-------CAGAAAAAUAUGCAAAAA ...((((((((((..........((((((....))))))....(((((...(((((((....))))))).(((((((....))))..)-------)))))))))).))))))) ( -31.90, z-score = -2.55, R) >droAna3.scaffold_13266 5402647 100 - 19884421 GGAUUUUUCCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGGC------GACGUGGCGAUAAAAAUCGAAC-------AAGAAAAAUAUGCAAAAA (((....)))..((((.......((((((....))))))....((((((((((.....))------)))((..((((....)))).))-------..)))))...)))).... ( -29.50, z-score = -3.04, R) >dp4.chr3 16872205 89 + 19779522 GGGCUCUGGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGAC-----------------GACGAUAAAAAUCGAGU-------AAGAAAAAUAUGCAAAGA .(((....))).((((.......((((((....))))))....(((((....((-----------------..((((....)))).))-------..)))))...)))).... ( -21.00, z-score = -1.56, R) >droPer1.super_2 5938211 89 + 9036312 GGGCUCUGGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGAC-----------------GACGAUAAAAAUCGAGC-------AAGAAAAAUAUGCAAAGA ..((((.................((((((....))))))...(((((..(((..-----------------..)))..))))).))))-------.................. ( -19.40, z-score = -0.80, R) >droVir3.scaffold_12875 13860673 92 + 20611582 -------------GCACAUAAAAGCGCAAAUAUUUGCGCAACAAUUUCA--AAUA---UCGUCACAGACGUUGCGAUAAAAAUCGAACAAG---CAAAAUAAAUAUGCAAAAU -------------(((.......((((((....))))))..........--..((---((((.((....)).)))))).............---...........)))..... ( -18.40, z-score = -2.41, R) >droMoj3.scaffold_6496 13470264 108 - 26866924 ACGCUUGGGAAAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAAUUUCA--AAUA---UCGUCGCAGACGUUGCGAUAAAAAUCGAACAUGCUACAACAAAAAUAUGCAAAAU ............((((.......((((((....))))))..........--....---..((.(((...(((.((((....))))))).))).))..........)))).... ( -21.50, z-score = -0.73, R) >droGri2.scaffold_15112 3548384 93 + 5172618 ------------UGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCA--AACAUCGUCGUCGCAGACGCGGCGAUAAAAAUCGAACA------AGAAAAUAUAUGCAAAAU ------------((((.(((...((((((....))))))..........--......((((((((....))))))))............------......))).)))).... ( -25.80, z-score = -3.47, R) >consensus AGGUUUUUGCUAUGCACAUAAAAGCGCAAAUAUUUGCGCAACAUUUUCGUCGUCGUCGUC______GAC__GGCGAUAAAAAUCGAAC_______AAGAAAAAUAUGCAAAAA ............((((.......((((((....))))))..................................((((....))))....................)))).... (-14.45 = -14.54 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:13 2011