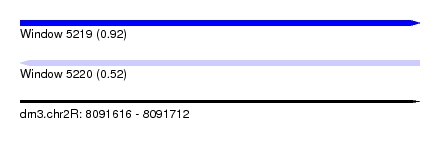
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,091,616 – 8,091,712 |
| Length | 96 |
| Max. P | 0.917975 |

| Location | 8,091,616 – 8,091,712 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.31485 |
| G+C content | 0.34033 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
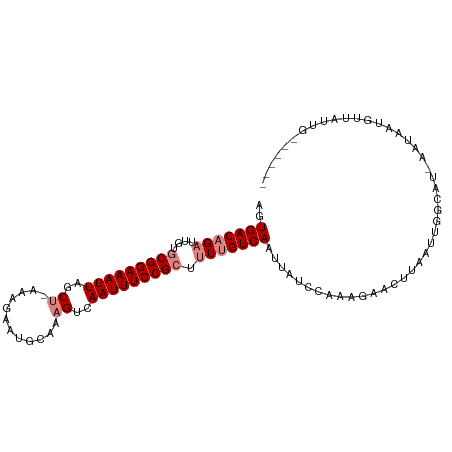
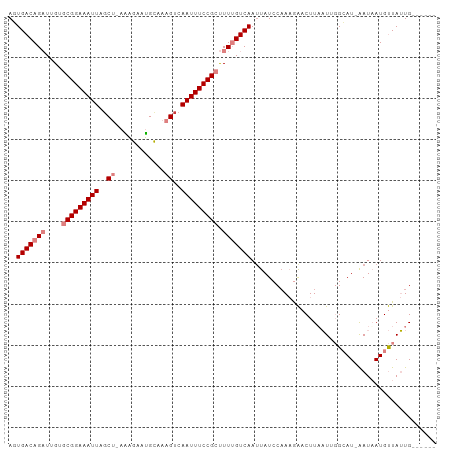
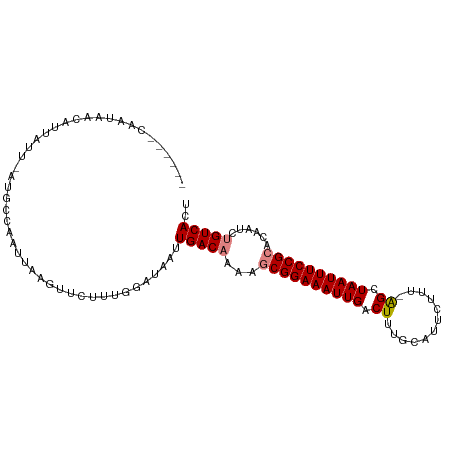
>dm3.chr2R 8091616 96 + 21146708 AUUGACAGAUUGUGCGGAAAUUAGCU-AAAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUAGGCAU-AAUAAUGUUAUUG------ (((((((((....(((((((((..((-...........))..))))))))).)))))))))..................(((((-....)))))....------ ( -23.60, z-score = -2.96, R) >droSim1.chr2R_random 2181733 96 + 2996586 AGUGACAGAUUGUGCGGAAAUUAGCU-AAAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUAGGCAU-AAUAAUGUUAUUG------ ..(((((((....(((((((((..((-...........))..))))))))).)))))))....................(((((-....)))))....------ ( -22.50, z-score = -2.46, R) >droSec1.super_1 5636186 96 + 14215200 AGUGACAGAUUGUGCGGAAAUUAGCU-AAAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUAGGCAU-AAUAAUGUUAUUG------ ..(((((((....(((((((((..((-...........))..))))))))).)))))))....................(((((-....)))))....------ ( -22.50, z-score = -2.46, R) >droYak2.chr2R 12280334 96 - 21139217 AGUGACAGAUUGUGCGGAAAUUAGCC-AGAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCUAAGAACUUUAUUGGCAU-AAUAAUGUUAUUG------ ..(((((((....(((((((((..(.-............)..))))))))).)))))))(((((.((((........)))).))-)))..........------ ( -22.42, z-score = -1.99, R) >droEre2.scaffold_4845 4885014 96 - 22589142 AGUGACAGAUUGUGCGGAAAUUAUCC-AAAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAAUAUCCAAAGAACUUUAUUGGCAU-AAUAAUGUUAUUG------ ..(((((((....(((((((((..(.-............)..))))))))).)))))))...................((((((-....))))))...------ ( -19.42, z-score = -1.48, R) >dp4.chr3 16856401 103 - 19779522 ACUGACAGCUUGAACGGAAAUUAGCUUAGGAAACGUAAAGUCAAUUUCCGGCUU-GUCAAUUAUACAACAGCCUUAUUUCGGAUUGAUUAUGCUAAUGCCGUAA ..(((((((.....((((((((..(((.(....)...)))..)))))))))).)-))))....(((..(((((.......)).))).....((....)).))). ( -24.20, z-score = -1.63, R) >droPer1.super_2 5922298 103 - 9036312 ACUGACAGCUUGAACGGAAAUUAGCUUAGGAAACGUAAAGUCAAUUUCCGGCUU-GUCAAUUAUACAACAGCCUUAUUUCGGAUUGAUUAUGCUAAUGCCGUAA ..(((((((.....((((((((..(((.(....)...)))..)))))))))).)-))))....(((..(((((.......)).))).....((....)).))). ( -24.20, z-score = -1.63, R) >consensus AGUGACAGAUUGUGCGGAAAUUAGCU_AAAGAAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUUGGCAU_AAUAAUGUUAUUG______ ..(((((((....(((((((((..((..(....)....))..))))))))).)))))))............................................. (-15.78 = -16.47 + 0.69)

| Location | 8,091,616 – 8,091,712 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.31485 |
| G+C content | 0.34033 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
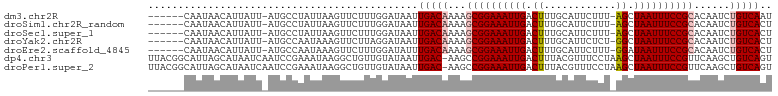
>dm3.chr2R 8091616 96 - 21146708 ------CAAUAACAUUAUU-AUGCCUAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUUCUUU-AGCUAAUUUCCGCACAAUCUGUCAAU ------...........((-((.((.............))))))((((((...((((((((((.((...........-)).))))))))))......)))))). ( -17.82, z-score = -1.64, R) >droSim1.chr2R_random 2181733 96 - 2996586 ------CAAUAACAUUAUU-AUGCCUAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUUCUUU-AGCUAAUUUCCGCACAAUCUGUCACU ------..........(((-((.((.............)))))))(((((...((((((((((.((...........-)).))))))))))......))))).. ( -17.22, z-score = -1.56, R) >droSec1.super_1 5636186 96 - 14215200 ------CAAUAACAUUAUU-AUGCCUAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUUCUUU-AGCUAAUUUCCGCACAAUCUGUCACU ------..........(((-((.((.............)))))))(((((...((((((((((.((...........-)).))))))))))......))))).. ( -17.22, z-score = -1.56, R) >droYak2.chr2R 12280334 96 + 21139217 ------CAAUAACAUUAUU-AUGCCAAUAAAGUUCUUAGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUUCUCU-GGCUAAUUUCCGCACAAUCUGUCACU ------..........(((-((.((.............)))))))(((((...((((((((((.((...........-)).))))))))))......))))).. ( -16.72, z-score = -1.05, R) >droEre2.scaffold_4845 4885014 96 + 22589142 ------CAAUAACAUUAUU-AUGCCAAUAAAGUUCUUUGGAUAUUUGACAAAAGCGGAAAUUGACUUUGCAUUCUUU-GGAUAAUUUCCGCACAAUCUGUCACU ------(((.(((.(((((-.....))))).)))..)))......(((((...((((((((((.((...........-)).))))))))))......))))).. ( -18.30, z-score = -1.55, R) >dp4.chr3 16856401 103 + 19779522 UUACGGCAUUAGCAUAAUCAAUCCGAAAUAAGGCUGUUGUAUAAUUGAC-AAGCCGGAAAUUGACUUUACGUUUCCUAAGCUAAUUUCCGUUCAAGCUGUCAGU ..(((((...(((..................((((((((......))))-.))))((((((((.(((..........))).)))))))))))...))))).... ( -23.30, z-score = -1.63, R) >droPer1.super_2 5922298 103 + 9036312 UUACGGCAUUAGCAUAAUCAAUCCGAAAUAAGGCUGUUGUAUAAUUGAC-AAGCCGGAAAUUGACUUUACGUUUCCUAAGCUAAUUUCCGUUCAAGCUGUCAGU ..(((((...(((..................((((((((......))))-.))))((((((((.(((..........))).)))))))))))...))))).... ( -23.30, z-score = -1.63, R) >consensus ______CAAUAACAUUAUU_AUGCCAAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUUCUUU_AGCUAAUUUCCGCACAAUCUGUCACU .............................................(((((...((((((((((.((............)).))))))))))......))))).. (-12.96 = -13.33 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:17:10 2011