
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,003,367 – 8,003,512 |
| Length | 145 |
| Max. P | 0.704742 |

| Location | 8,003,367 – 8,003,512 |
|---|---|
| Length | 145 |
| Sequences | 6 |
| Columns | 145 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.45067 |
| G+C content | 0.45307 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -15.04 |
| Energy contribution | -20.35 |
| Covariance contribution | 5.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
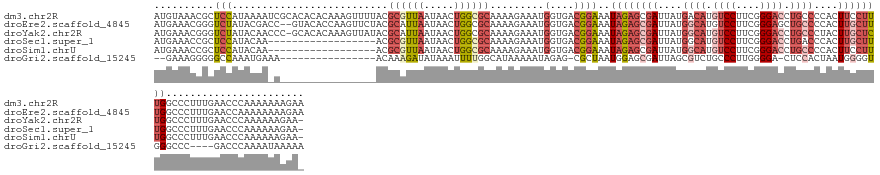
>dm3.chr2R 8003367 145 + 21146708 AUGUAAACGCUCCAUAAAAUCGCACACACAAAGUUUUACGCGUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGACAUGUCCUUCGGGACCUGCCCCACUUCCUUUGGCCCUUUGAACCCAAAAAAAGAA ..((((.(((((.((....((((((.((.....((((..((((((.....))))))..))))..))))).)))..)).))))).))))..((.((((....)))).)).....(((..(((((..........)))))..))).. ( -32.10, z-score = -0.62, R) >droEre2.scaffold_4845 4797733 143 - 22589142 AUGAAACGGGUCUAUACGACC--GUACACCAAGUUCUACGCAUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGGCAUGUCCUUCGGGAGCUGCCCCACUUGCUUUGGCCCUUUGAACCAAAAAAAAGAA .......(((((.......((--((.(((((..((((.(((...........)))....)))).)))))))))....(((((((....((((..(((....)))..))))....))))))))))))................... ( -44.20, z-score = -2.92, R) >droYak2.chr2R 12191193 143 - 21139217 AUGAAACGGGUCUAUACAACCC-GCACACAAAGUUAUACGCAUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGGCAUGUCCUUCGGGACCUGCCCUACUUGCUCUGGCCCUUUGAACCCAAAAAAGAA- .......((((...........-((.(....((((((........)))))).).)).((((....(....)((...((((((((.((.((((.((((....)))).)))).)).)))))))).))))))..)))).........- ( -45.50, z-score = -3.78, R) >droSec1.super_1 5551711 126 + 14215200 AUGAAACCGCUCCAUACAA------------------ACGCGUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGGCAUGUCCUUCGGGACCUGACCCACUUGCUUUGGCCCUUUGAACCCAAAAAAGAA- ......((((.((((....------------------..((((((.....)))))).......)))).).)))...((((((((....((((.((((....)))).)).))...))))))))...((((.........))))..- ( -34.02, z-score = -2.05, R) >droSim1.chrU 6015547 126 - 15797150 AUGAAACCGCUCCAUACAA------------------ACGCGUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGGCAUGUCCUUCGGGACCUGCCCCACUUCCUUUGGCCCUUUGAACCCAAAAAAGAA- .......(((((.......------------------..((((((.....))))))........((....))......))))).....((((.((((....)))).))))...(((..(((((..........))))).)))..- ( -35.00, z-score = -2.17, R) >droGri2.scaffold_15245 16435495 121 - 18325388 --GAAAGGGGGCCAAAUGAAA----------------ACAAAGAUAUAAAUUUUGGCAUAAAAAUAGAG-CGCUAAUGGAGCGAUUAGCGUCUGCCCUUGGGGA-CUCCACUAAUGGGGUGGGCCC----GACCCAAAAUAAAAA --..(((((.((((((.....----------------..............)))))).......((((.-(((((((......)))))))))))))))).(((.-(.(((((.....)))))))))----............... ( -36.01, z-score = -1.66, R) >consensus AUGAAACCGCUCCAUACAA_C________________ACGCAUUAAUAACUGGCGCAAAAGAAAUGGUGACGGAAAUAGAGCGAUUAUGGCAUGUCCUUCGGGACCUGCCCCACUUGCUUUGGCCCUUUGAACCCAAAAAAAAA_ .......................................((((((.....)))))).........(((...((...((((((((....((((.((((....)))).))))....))))))))..)).....)))........... (-15.04 = -20.35 + 5.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:58 2011