
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,996,612 – 7,996,712 |
| Length | 100 |
| Max. P | 0.693425 |

| Location | 7,996,612 – 7,996,712 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.04 |
| Shannon entropy | 0.62597 |
| G+C content | 0.44256 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -10.64 |
| Energy contribution | -9.12 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7996612 100 + 21146708 CUCUGGCAUUUCCCAGCUAGCAAUUUUCACGUUUUUCCCGAAACUGUUUGCUUUGCUUUCUGCCAA--CUGGCUUUUGCAUACUUCACUGCU--AUGCA--------ACUCU---- ...(((((......(((.(((((....((.(((((....))))))).)))))..)))...))))).--.......((((((((......).)--)))))--------)....---- ( -21.70, z-score = -2.22, R) >droEre2.scaffold_4845 4791036 96 - 22589142 CUCUGGCAUUUUCCAGCUCCCAAUUUUCACGUUUU-CCCGGAACUGUUUGCUCUGCUUUCUGCCCC--UGGGCUUUUGCAUUC-------------GCAGAGGAACAACUCC---- ..((((......))))...................-...(((..(((((.((((((.....(((..--..)))..........-------------)))))))))))..)))---- ( -22.16, z-score = -0.51, R) >droYak2.chr2R 12184376 96 - 21139217 CUCUGGCUUUUCCCAGCUCCCAAUUUUCACGUUUU-CCCGAAACUGUUUGCCUUGCUUGCUGCCAC--CUGGCUUUUGCAUAC-------------GCAGAGAAAAAACUCU---- ..((((......)))).......(((((..(((((-...)))))..(((((..(((.....(((..--..)))....)))...-------------))))))))))......---- ( -19.40, z-score = -0.80, R) >droSec1.super_1 5545071 110 + 14215200 CUCUGGCAUUUUCCAGCUAGCAAUUUUCACGUUUUUCCCGGAACUGUUUGCUUUGCUUUCUGCCAA--CUGGCUUUUGCAUACUUCAAUGUUUUAUGCAGAGAGAAAACUCU---- ((((((((......(((.(((((....((.((((......)))))).)))))..)))...))))).--......((((((((...........)))))))))))........---- ( -23.80, z-score = -0.56, R) >droSim1.chr2R 6528201 110 + 19596830 CUCUGGCAUUUUCCAGCUAGCAAUUUUCACGUUUUUCCCGAAACUGUUUGCUUUGCUUUCUGCCAA--CUGGCUUUUGCAUACUUCAAUGUUUUAUGCAGAGAGAAAACUCU---- ((((((((......(((.(((((....((.(((((....))))))).)))))..)))...))))).--......((((((((...........)))))))))))........---- ( -24.00, z-score = -1.04, R) >droMoj3.scaffold_6496 18743639 108 + 26866924 ----GCAAAGCUGCGACUGCCAACGAGCACAUAUUCUCUAGUAUU-UUCAUUUGGUAUUUUUAUAAUACUAACCACUGACUGAUUCCACUUCA---ACGGGCUAACCACACGCCAA ----((...(((.((........)))))..........((((...-.....((((((((.....))))))))...(((..(((.......)))---.))))))).......))... ( -14.70, z-score = 0.19, R) >consensus CUCUGGCAUUUUCCAGCUACCAAUUUUCACGUUUUUCCCGAAACUGUUUGCUUUGCUUUCUGCCAA__CUGGCUUUUGCAUACUUCAAUGUU____GCAGAGAAAAAACUCU____ ..((((......))))..............(((((....)))))..(((((..(((.....((((....))))....)))................)))))............... (-10.64 = -9.12 + -1.52)

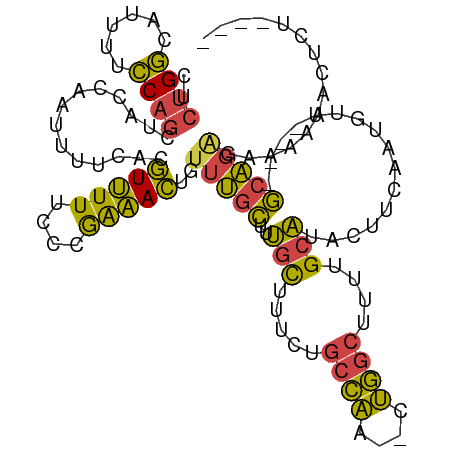

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:57 2011