
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,953,746 – 7,953,857 |
| Length | 111 |
| Max. P | 0.996345 |

| Location | 7,953,746 – 7,953,857 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.60 |
| Shannon entropy | 0.46459 |
| G+C content | 0.37157 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
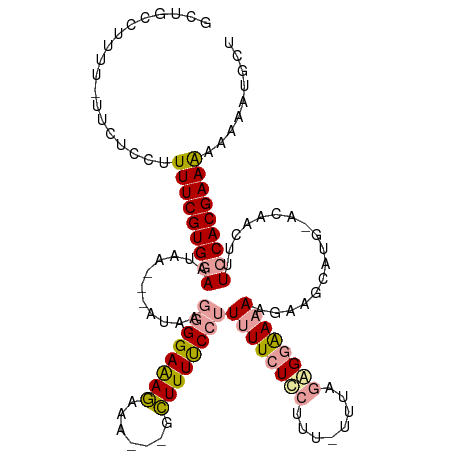
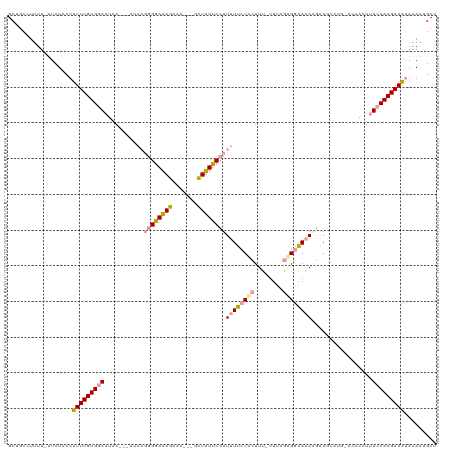
>dm3.chr2R 7953746 111 + 21146708 GCUGCCUUUU-UUCUUCUUUUCGUGGAAUAAUGUAUAAGGGGAAAGAAAAGAGCUUUUCCUUUUUCUCAUUU-UUUUAAGUGAAAGUGA-------CAACUUUCCACGAAAAAAAGUGCU ...(((((((-((.......((((((((...(((..((((((((((.......))))))))))....(((((-((......))))))))-------))...))))))))))))))).)). ( -31.01, z-score = -2.71, R) >droSim1.chr3L 7848602 120 + 22553184 GCUCCGUUUUAUUCUCCUUUUCGUGAAAUAAGUAAUAAGGGGAAAAAAAGAGCUUUUCCCUUUUUCUGCUUUGUUUCGUGGAAUACGAAGCAUGUACAACUCUCCACGAAGCAAAAAGCU ((((..((((.((((((((.................)))))))).))))))))..............(((((((((((((((....................)))))))))))..)))). ( -31.68, z-score = -2.56, R) >droSec1.super_1 5503064 103 + 14215200 GCUGCUUAUU-GUCUCCUUUUCGUGGAAUAA---------GGAGAG------GCUUUUCCUUUUUCUCCUUU-UUUAGGGGAAAAAGAAGCAUGCACAACUUUCCACGAAAAAAAUUGCC ..........-......(((((((((((...---------.(.(..------((((...(((((((((((..-...)))))))))))))))...).)....)))))))))))........ ( -32.10, z-score = -2.70, R) >consensus GCUGCCUUUU_UUCUCCUUUUCGUGGAAUAA___AUAAGGGGAAAGAAA___GCUUUUCCUUUUUCUCCUUU_UUUAGAGGAAAAAGAAGCAUG_ACAACUUUCCACGAAAAAAAAUGCU ..................(((((((((...........((((((((.......))))))))((((((((........)))))))).................)))))))))......... (-18.84 = -19.63 + 0.79)

| Location | 7,953,746 – 7,953,857 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.60 |
| Shannon entropy | 0.46459 |
| G+C content | 0.37157 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
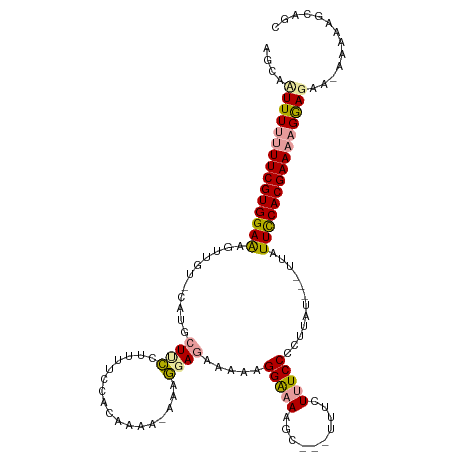
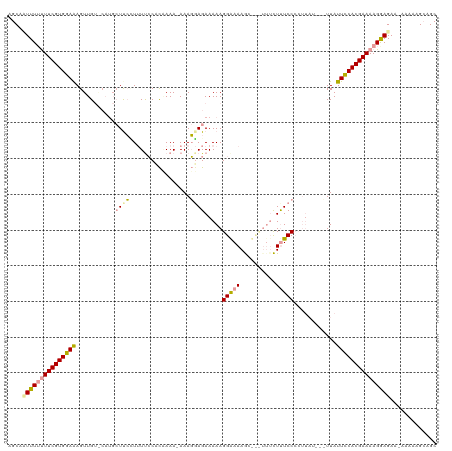
>dm3.chr2R 7953746 111 - 21146708 AGCACUUUUUUUCGUGGAAAGUUG-------UCACUUUCACUUAAAA-AAAUGAGAAAAAGGAAAAGCUCUUUUCUUUCCCCUUAUACAUUAUUCCACGAAAAGAAGAA-AAAAGGCAGC ....((((((((((((((((((..-------..))))))........-..(((((...((((((((....))))))))...))))).........))))))))))))..-.......... ( -27.60, z-score = -2.61, R) >droSim1.chr3L 7848602 120 - 22553184 AGCUUUUUGCUUCGUGGAGAGUUGUACAUGCUUCGUAUUCCACGAAACAAAGCAGAAAAAGGGAAAAGCUCUUUUUUUCCCCUUAUUACUUAUUUCACGAAAAGGAGAAUAAAACGGAGC ...(((((((((((((((((((.......)))).....))))))).....))))))))..((((((((.....))))))))(((.....((((((..(.....)..))))))....))). ( -28.50, z-score = -0.98, R) >droSec1.super_1 5503064 103 - 14215200 GGCAAUUUUUUUCGUGGAAAGUUGUGCAUGCUUCUUUUUCCCCUAAA-AAAGGAGAAAAAGGAAAAGC------CUCUCC---------UUAUUCCACGAAAAGGAGAC-AAUAAGCAGC .((.....(((((((((((((..(.((.(..(((((((((.(((...-..))).))))))))).).))------..)..)---------)..)))))))))))(....)-.....))... ( -31.10, z-score = -2.64, R) >consensus AGCAAUUUUUUUCGUGGAAAGUUGU_CAUGCUUCCUUUUCCACAAAA_AAAGGAGAAAAAGGAAAAGC___UUUCUUUCCCCUUAU___UUAUUCCACGAAAAGGAGAA_AAAAAGCAGC ....(((((((((((((((...........((((.................)))).....(((((..........)))))............)))))))))))))))............. (-16.24 = -16.70 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:49 2011