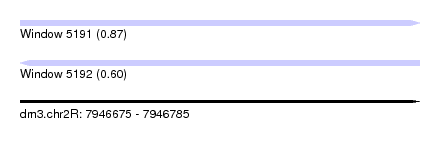
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,946,675 – 7,946,785 |
| Length | 110 |
| Max. P | 0.871937 |

| Location | 7,946,675 – 7,946,785 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.36579 |
| G+C content | 0.45644 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
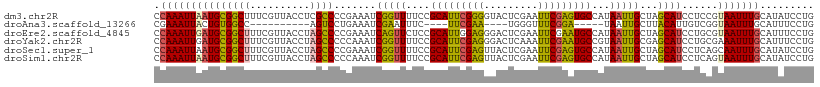
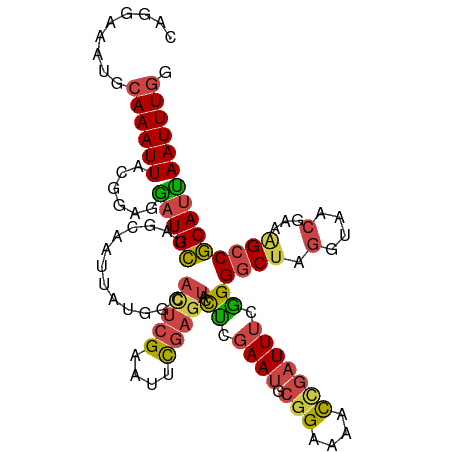
>dm3.chr2R 7946675 110 + 21146708 CCAAAUUAAUGCGGCUUUCGUUACCUCGCCCCGAAAUCGGUUUUCCGCAUUCGGGGUACUCGAAUUCGAGUGCCAUAAUUGCUAGCAUCCUCCGUAAUUUGCAUAUCCUG ........(((((((............((((((((..(((....)))..))))))))(((((....))))))))..((((((...........)))))).))))...... ( -34.20, z-score = -3.64, R) >droAna3.scaffold_13266 17204119 87 - 19884421 CGAAAUUACUGUGGCC----------AGUCCUGAAAUCGAAUUUC----UUCGAA----UGGGUUUCGGA-----UAAUUGCUUACAUUGUCGGUAAUUUGCAUUUCCUG ..(((((((((.(((.----------.((((.((((((..((((.----...)))----).)))))))))-----)....)))........))))))))).......... ( -17.50, z-score = -0.29, R) >droEre2.scaffold_4845 4739071 110 - 22589142 CCAAAUUGAUGCGGCUUUCGUUACCUAGCCCCGAAAUCAGUUCUCCGCAUUGGAGGGACUCGAAUUCGAAUGCCAUAAUUGCUAGCAUCCUGCGUAAUUUGCAUUUCCUG .......(((((((((..........)))).((((.((((((((((.....))).))))).)).))))................))))).((((.....))))....... ( -26.60, z-score = -0.68, R) >droYak2.chr2R 12133313 110 - 21139217 CCAAAUUGAUGCGGCUUUCGUUACCUAGCCCCCAAAUCGGUUUUCCGCAUUCGAGGGACUCAAAUUCGAAUGCCGUAAUUGCGAGCAUCCUGCGAAAUUUGCAUUUCCUG .((((((..((((((.((((......(((((.(.((((((....))).))).).))).))......)))).)))))).(((((.......)))))))))))......... ( -29.40, z-score = -1.41, R) >droSec1.super_1 5496052 110 + 14215200 CCAAAUUAAUGCGGCUUUCGUUACCUAGCCCCGAAAUCGGUUUUCCGCAUUCGAGUUACUCGAAUUCGAGUGCCAUAAUUGCUAGCAUCCUCAGCAAUUUGCAUAUCCUG ........((((((((..........))))(((....)))......(((((((((((.....)))))))))))...(((((((((....)).))))))).))))...... ( -29.40, z-score = -3.32, R) >droSim1.chr2R 6483756 110 + 19596830 CCAAAUUAAUGCGGCUUUCGUUACCUAGCCCCCAAAUCGGUUUUCCGCAUUCGAGUUACUCGAAUUCGAGUGCCAUAAUUGCUAGCAUCCUCAGUAAUUUGCAUAUCCUG .(((((((.((.((..........(((((.........((....))(((((((((((.....))))))))))).......)))))...)).)).)))))))......... ( -25.82, z-score = -2.94, R) >consensus CCAAAUUAAUGCGGCUUUCGUUACCUAGCCCCGAAAUCGGUUUUCCGCAUUCGAGGUACUCGAAUUCGAAUGCCAUAAUUGCUAGCAUCCUCCGUAAUUUGCAUAUCCUG .(((((((((((((((..........)))).......(((((....(((((((((.........)))))))))...)))))...))))......)))))))......... (-16.42 = -16.43 + 0.02)
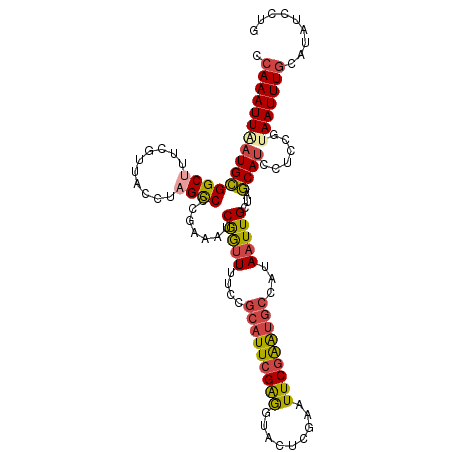

| Location | 7,946,675 – 7,946,785 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.36579 |
| G+C content | 0.45644 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
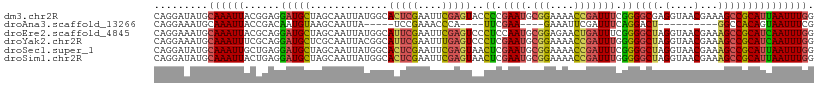
>dm3.chr2R 7946675 110 - 21146708 CAGGAUAUGCAAAUUACGGAGGAUGCUAGCAAUUAUGGCACUCGAAUUCGAGUACCCCGAAUGCGGAAAACCGAUUUCGGGGCGAGGUAACGAAAGCCGCAUUAAUUUGG .........((((((.(((.((..((((.......))))(((((....))))).)))))(((((((....(((....)))..........(....)))))))))))))). ( -34.80, z-score = -2.57, R) >droAna3.scaffold_13266 17204119 87 + 19884421 CAGGAAAUGCAAAUUACCGACAAUGUAAGCAAUUA-----UCCGAAACCCA----UUCGAA----GAAAUUCGAUUUCAGGACU----------GGCCACAGUAAUUUCG ..(((..(((...((((.......)))))))....-----)))((((((..----.(((((----....))))).....))(((----------(....))))..)))). ( -17.20, z-score = -1.65, R) >droEre2.scaffold_4845 4739071 110 + 22589142 CAGGAAAUGCAAAUUACGCAGGAUGCUAGCAAUUAUGGCAUUCGAAUUCGAGUCCCUCCAAUGCGGAGAACUGAUUUCGGGGCUAGGUAACGAAAGCCGCAUCAAUUUGG ((((...(((.......))).(((((.........((((.((((((.((.(((..((((.....)))).))))).))))))))))(((.......))))))))..)))). ( -30.20, z-score = -0.87, R) >droYak2.chr2R 12133313 110 + 21139217 CAGGAAAUGCAAAUUUCGCAGGAUGCUCGCAAUUACGGCAUUCGAAUUUGAGUCCCUCGAAUGCGGAAAACCGAUUUGGGGGCUAGGUAACGAAAGCCGCAUCAAUUUGG ...(((((....)))))....(((((..(......)(((.((((......((((((.(((((.(((....))))))))))))))......)))).))))))))....... ( -35.80, z-score = -2.12, R) >droSec1.super_1 5496052 110 - 14215200 CAGGAUAUGCAAAUUGCUGAGGAUGCUAGCAAUUAUGGCACUCGAAUUCGAGUAACUCGAAUGCGGAAAACCGAUUUCGGGGCUAGGUAACGAAAGCCGCAUUAAUUUGG ((((..((((.((((((((.......)))))))).(((((((((....)))))..((((((..(((....)))..))))))))))(((.......)))))))...)))). ( -35.10, z-score = -3.14, R) >droSim1.chr2R 6483756 110 - 19596830 CAGGAUAUGCAAAUUACUGAGGAUGCUAGCAAUUAUGGCACUCGAAUUCGAGUAACUCGAAUGCGGAAAACCGAUUUGGGGGCUAGGUAACGAAAGCCGCAUUAAUUUGG .........(((((((.((.((...(((((.........(((((....)))))..(((((((.(((....)))))))))).)))))....(....))).)).))))))). ( -33.20, z-score = -3.23, R) >consensus CAGGAAAUGCAAAUUACGGAGGAUGCUAGCAAUUAUGGCACUCGAAUUCGAGUACCUCGAAUGCGGAAAACCGAUUUCGGGGCUAGGUAACGAAAGCCGCAUUAAUUUGG .........((((((......(((((.............(((((....)))))..((.((((.(((....))))))).))((((..........))))))))))))))). (-13.82 = -13.98 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:46 2011