
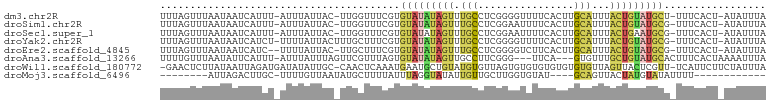
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,937,584 – 7,937,681 |
| Length | 97 |
| Max. P | 0.759933 |

| Location | 7,937,584 – 7,937,681 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 66.59 |
| Shannon entropy | 0.70196 |
| G+C content | 0.28487 |
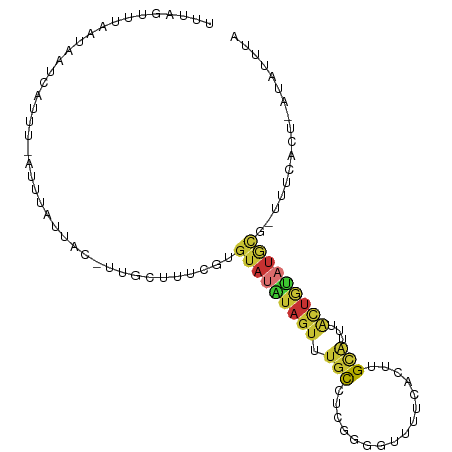
| Mean single sequence MFE | -14.97 |
| Consensus MFE | -5.35 |
| Energy contribution | -5.54 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.93 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759933 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 7937584 97 - 21146708 UUUAGUUUAAUAAUCAUUU-AUUUAUUAC-UUGGUUUCGUGUAUAUAGUUUGCCUCGGGGUUUUCACUUGCAUUUACUGUAUGCU-UUUCACU-AUAUUUA .......((((((......-..)))))).-.((((.....(((((((((.(((...((........)).)))...))))))))).-....)))-)...... ( -15.20, z-score = -1.69, R) >droSim1.chr2R 6474780 97 - 19596830 UUUAGUUUAAUAAUCAUUU-AUUUAUUAC-UUGGUUUCGUGUAUAUAGUUUGCCUCGGAAUUUUCACUUGCAUUUACUGUAUGCG-UUUCACU-AUAUUUA ..((((..((((((((..(-(.....)).-.)))))....(((((((((.(((....((....))....)))...))))))))))-))..)))-)...... ( -16.30, z-score = -2.16, R) >droSec1.super_1 5487453 97 - 14215200 UUUAGUUUAAUAAUCAUUU-AUUUAUUAC-UUGGUUUCGUGUAUAUAGUUUGCCUCGGAAUUUUCACUUGCAUUUACUGAAUGCG-UUUCACU-AUAUUUA ..((((..((((((((..(-(.....)).-.)))))....((((.((((.(((....((....))....)))...)))).)))))-))..)))-)...... ( -11.90, z-score = -0.22, R) >droYak2.chr2R 12124216 98 + 21139217 UUUAGUUUAAUAAUCAUCU-UUUUAUUACUUUGCUUUCGUGUAUAUAGUUUGCCUCGGGGUUUUCACUUGCAUUUACUGUAUGCG-UUUCACU-AUAUUUA ..((((.((((((......-..))))))............(((((((((.(((...((........)).)))...))))))))).-....)))-)...... ( -15.00, z-score = -1.72, R) >droEre2.scaffold_4845 4730450 96 + 22589142 UUUAGUUUAAUAAUCAUC--UUUUAUUAC-UUGCUUUCGUGUAUAUAGUUUGCCUCGGGGUCUUCACUUGCAUUUACUGUAUGCG-UUUCACU-AUAUUUA ..((((.((((((.....--..)))))).-..........(((((((((.(((...((........)).)))...))))))))).-....)))-)...... ( -14.70, z-score = -1.39, R) >droAna3.scaffold_13266 17195970 94 + 19884421 UUUUGUUUAAUAUUCAUUU-AUUUAUUUAGUUCGUUUAGUGUAUAUAGUUGCCUUCGGG---UUCA---GUGUUUGCUGUAUGCACUUUCACUAAAAUUUA ...................-.....((((((......((((((((((((.(((....))---)...---......))))))))))))...))))))..... ( -22.20, z-score = -4.70, R) >droWil1.scaffold_180772 6206800 98 + 8906247 -GAACUCUUAUAAUUAGAUGAUAUAUUGC-CAACUCAAAUGAAUGCUGUAUGUGUUAGUGUGUGUGUGUGUGUUAGUUACUCGUU-UCAUUCUUCUAUUUA -(((((((.......)))((((((((...-..((.((....(((((.....)))))..)).))....)))))))).......)))-).............. ( -10.40, z-score = 1.95, R) >droMoj3.scaffold_6496 18644026 76 - 26866924 --------AUUAGACUUGC-UUUUGUUAAUAUGCUUUUAUUUAGGUAUAUUGUUGCUUGGUGUAU----GCAGUUACUAUGUAUAUUUU------------ --------.....(((.((-......((((((((((......))))))))))..))..)))((((----(((.......)))))))...------------ ( -14.10, z-score = -1.07, R) >consensus UUUAGUUUAAUAAUCAUUU_AUUUAUUAC_UUGCUUUCGUGUAUAUAGUUUGCCUCGGGGUUUUCACUUGCAUUUACUGUAUGCG_UUUCACU_AUAUUUA ........................................(((((((((.(((....((....))....)))...)))))))))................. ( -5.35 = -5.54 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:43 2011