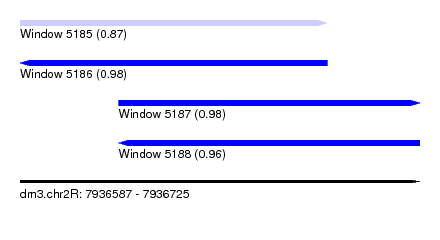
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,936,587 – 7,936,725 |
| Length | 138 |
| Max. P | 0.980189 |

| Location | 7,936,587 – 7,936,693 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.96 |
| Shannon entropy | 0.64208 |
| G+C content | 0.56650 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.65 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
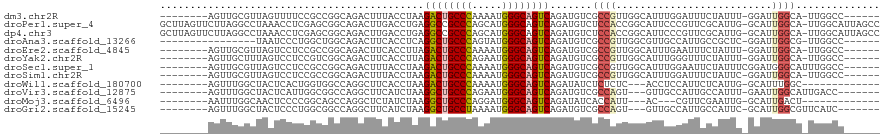
>dm3.chr2R 7936587 106 + 21146708 -UGGGAUCCUCCGCUG--CCGAAGCGGCUGCCAAUGAGGCCAA-UGCCAAUCCAAA------UAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAG -..((((..((.(((.--....)))(((.(((.....)))...-.)))........------..)).))))...((((...))))......((((((((.....)))))))).... ( -35.30, z-score = -1.64, R) >droPer1.super_4 5326229 106 - 7162766 -------UCUCCUCGG--UGGAGGCGGCGGCCAACGAGGCUAA-UGCCAAUGCCAAUGCCAAUGCGAACGGGAAUGCCGGUGGAGACAUCUGACUGCCCAUGCUGGGCGGCCUCAG -------(((((((((--(...((((..((((.....))))..-))))....((..(((....)))....))...))))).)))))...((((((((((.....))))))..)))) ( -42.30, z-score = -0.51, R) >dp4.chr3 11854377 106 - 19779522 -------UCUCCUCGG--UGGAGGCGGCGGCCAACGAGGCUAA-UGCCAAUGCCAAUGCCAAUGCGAACGGGAAUGCCGGUGGAGACAUCUGACUGCCCAUGCUGGGCGGCCUCAG -------(((((((((--(...((((..((((.....))))..-))))....((..(((....)))....))...))))).)))))...((((((((((.....))))))..)))) ( -42.30, z-score = -0.51, R) >droAna3.scaffold_13266 17194905 106 - 19884421 ---UGGUGACCAGCGCUGCCGAGGUGGCCGCCAACGAGGCCAA-CGCCAAUCCGAG------CGGCAAUGGCAACGCCAACGGCGACAUCUGACUGCCCAUACUGGGCAGCCUGAG ---.((((.(((..(((((((.((((((.(((.....)))...-.))).))))).)------))))..)))...((((...)))).))))...((((((.....))))))...... ( -43.60, z-score = -1.23, R) >droEre2.scaffold_4845 4729442 106 - 22589142 -UGGGAUCCUCCGCUG--CCGAAGCGGCGGCCAAUGAGGCCAA-UGCCAAUCCAAA------UAGAAAUUCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUAAG -..((((...((((((--(....))))))).......(((...-.))).))))...------............((((...))))......((((((((.....)))))))).... ( -38.70, z-score = -2.39, R) >droYak2.chr2R 12123219 106 - 21139217 -UGGGAUCCUCCGCUG--CCGAAGCGGCGGCCAAUGAGGCCAA-UGCCAAUCCAAA------UAGAAACCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUCUGGGCAGUCUAAG -((((.....((((((--(....))))))).......(((...-.)))........------......))))..((((...))))......((((((((.....)))))))).... ( -40.90, z-score = -3.08, R) >droSec1.super_1 5486452 111 + 14215200 UGGGGAUCCUCCGCUUGCCCGAAGCGGCGGCCAAUGAGGCCAAAUGCCCAUCCGAAA-----UAGAAUUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAG ((((.((...((((((.....)))))).((((.....))))..)).)))).......-----............((((...))))......((((((((.....)))))))).... ( -41.00, z-score = -2.00, R) >droSim1.chr2R 6473782 106 + 19596830 -UGGGAUCCUCCGCUG--CCGAAGCGGCGGCCAAUGAGGCCAA-UGCCAAUCCGAA------UAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAG -..((((..((.((((--(....)))))((((.....))))..-............------..)).))))...((((...))))......((((((((.....)))))))).... ( -38.90, z-score = -2.10, R) >droVir3.scaffold_12875 12043020 93 - 20611582 -------------UGAAGCGGCUGUCGAGGUCAAUGAGGUCAA-UGCCAAUUCAAA------UGGCAAUGGCAACAC---UGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAG -------------.....(((.((((...((((.((..((((.-(((((.......------))))).))))..)).---)))))))).))).((((((.....))))))...... ( -34.70, z-score = -2.25, R) >droMoj3.scaffold_6496 18641501 84 + 26866924 -------------UGAGGCGUCUUCUCAAAACAACGAAGUCAA-UGCCAAUUCGAA------CGGUAA------------UGGUGAUAUCUGACUGCCCAUCCUGGGCAGCCUUAG -------------((((((................((.((((.-((((........------.)))).------------...)))).))....(((((.....))))))))))). ( -23.80, z-score = -1.69, R) >droGri2.scaffold_15245 16349399 99 - 18325388 -------UGAGAGCACAGCUGCCAAGGAGGCCAAUGAGAUGAA-CGCCAAUGCGAA------UGGCAAUGGCAACAC---UGGCGACAUCUGACUGCCCAUUUUAGGCAGCCUUAG -------..........((((((.((((((.((.(.(((((..-(((((.(((...------..))).((....)).---))))).))))).).)).)).)))).))))))..... ( -34.70, z-score = -2.41, R) >consensus ____G_UCCUCCGCGG__CCGAAGCGGCGGCCAAUGAGGCCAA_UGCCAAUCCGAA______UGGAAAUGCAAAUGCCAACGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAG ...................................(((((....((((.................................)))).........(((((.....)))))))))).. (-13.78 = -13.65 + -0.14)


| Location | 7,936,587 – 7,936,693 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.96 |
| Shannon entropy | 0.64208 |
| G+C content | 0.56650 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.49 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7936587 106 - 21146708 CUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUA------UUUGGAUUGGCA-UUGGCCUCAUUGGCAGCCGCUUCGG--CAGCGGAGGAUCCCA- ....((((((((.....))))))))....((.((((..((((((..(((....)------))..))).(((.-...(((.....))).)))))).)))--).))((......)).- ( -43.50, z-score = -2.22, R) >droPer1.super_4 5326229 106 + 7162766 CUGAGGCCGCCCAGCAUGGGCAGUCAGAUGUCUCCACCGGCAUUCCCGUUCGCAUUGGCAUUGGCAUUGGCA-UUAGCCUCGUUGGCCGCCGCCUCCA--CCGAGGAGA------- ....(((.((((.....)))).))).....(((((..(((.....))).(((....(((..((((...(((.-...)))......))))..)))....--.))))))))------- ( -36.90, z-score = 0.63, R) >dp4.chr3 11854377 106 + 19779522 CUGAGGCCGCCCAGCAUGGGCAGUCAGAUGUCUCCACCGGCAUUCCCGUUCGCAUUGGCAUUGGCAUUGGCA-UUAGCCUCGUUGGCCGCCGCCUCCA--CCGAGGAGA------- ....(((.((((.....)))).))).....(((((..(((.....))).(((....(((..((((...(((.-...)))......))))..)))....--.))))))))------- ( -36.90, z-score = 0.63, R) >droAna3.scaffold_13266 17194905 106 + 19884421 CUCAGGCUGCCCAGUAUGGGCAGUCAGAUGUCGCCGUUGGCGUUGCCAUUGCCG------CUCGGAUUGGCG-UUGGCCUCGUUGGCGGCCACCUCGGCAGCGCUGGUCACCA--- ....((((((((.....)))))))).((.((.(((((..(((..((((.(((((------.......)))))-.))))..)))..))))).)).))((..((....))..)).--- ( -53.10, z-score = -2.16, R) >droEre2.scaffold_4845 4729442 106 + 22589142 CUUAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGAAUUUCUA------UUUGGAUUGGCA-UUGGCCUCAUUGGCCGCCGCUUCGG--CAGCGGAGGAUCCCA- ....((((((((.....)))))))).(((.((.(((((((((((..(((....)------))..))).....-..((((.....)))))))((....)--)))))).)))))...- ( -46.10, z-score = -3.15, R) >droYak2.chr2R 12123219 106 + 21139217 CUUAGACUGCCCAGAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGGUUUCUA------UUUGGAUUGGCA-UUGGCCUCAUUGGCCGCCGCUUCGG--CAGCGGAGGAUCCCA- ....((((((((.....))))))))((((((((....))))))))(((.((((.------........(((.-...))).......((((.((....)--).)))))))).))).- ( -47.00, z-score = -2.70, R) >droSec1.super_1 5486452 111 - 14215200 CUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAAUUCUA-----UUUCGGAUGGGCAUUUGGCCUCAUUGGCCGCCGCUUCGGGCAAGCGGAGGAUCCCCA ....((((((((.....))))))))((((((((((...)))(((((((((...)-----)))))))).)))))))((((.....)))).((((((.....)))))).((....)). ( -47.10, z-score = -2.19, R) >droSim1.chr2R 6473782 106 - 19596830 CUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUA------UUCGGAUUGGCA-UUGGCCUCAUUGGCCGCCGCUUCGG--CAGCGGAGGAUCCCA- ....((((((((.....)))))))).(((.((.((((((((((((((((....)------))))))).....-..((((.....)))))))((....)--)))))).)))))...- ( -47.00, z-score = -2.98, R) >droVir3.scaffold_12875 12043020 93 + 20611582 CUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCA---GUGUUGCCAUUGCCA------UUUGAAUUGGCA-UUGACCUCAUUGACCUCGACAGCCGCUUCA------------- ....((((((((.....))))))))...(((((.((---(((..(.((.(((((------.......)))))-.)).)..)))))....))))).........------------- ( -35.30, z-score = -2.85, R) >droMoj3.scaffold_6496 18641501 84 - 26866924 CUAAGGCUGCCCAGGAUGGGCAGUCAGAUAUCACCA------------UUACCG------UUCGAAUUGGCA-UUGACUUCGUUGUUUUGAGAAGACGCCUCA------------- ....((((((((.....))))))))...........------------......------........(((.-((...((((......))))..)).)))...------------- ( -21.90, z-score = -0.46, R) >droGri2.scaffold_15245 16349399 99 + 18325388 CUAAGGCUGCCUAAAAUGGGCAGUCAGAUGUCGCCA---GUGUUGCCAUUGCCA------UUCGCAUUGGCG-UUCAUCUCAUUGGCCUCCUUGGCAGCUGUGCUCUCA------- ....(((((((......((.((((.(((((.(((((---((((.((....))..------...)))))))))-..))))).)))).)).....))))))).........------- ( -41.20, z-score = -3.45, R) >consensus CUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUCCAUUUCCA______UUCGGAUUGGCA_UUGGCCUCAUUGGCCGCCGCUUCGG__CAGCGGAGGA_C____ ....((((((((.....)))))))).(((((........)))))................................(((.....)))............................. (-18.40 = -18.49 + 0.09)
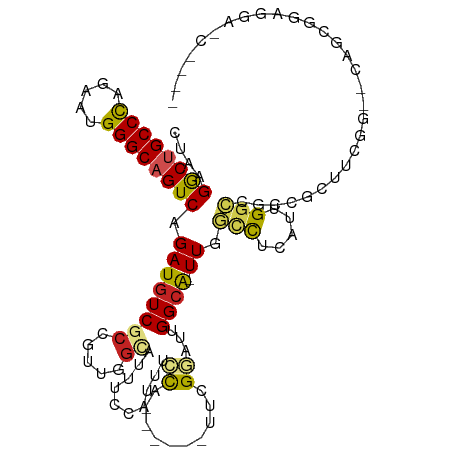
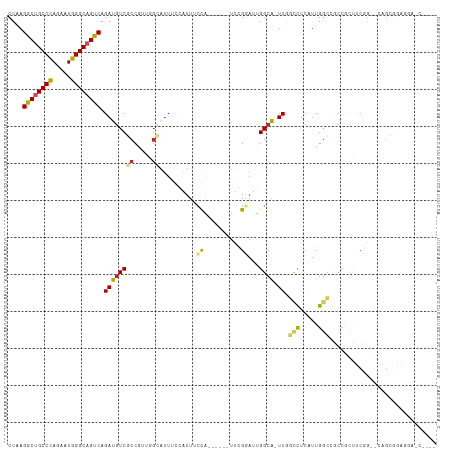
| Location | 7,936,621 – 7,936,725 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.62154 |
| G+C content | 0.53506 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -14.65 |
| Energy contribution | -13.38 |
| Covariance contribution | -1.26 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7936621 104 + 21146708 ------GGCCAA-UGCCAAUCC-AAAUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAAAACUAACGCAACU-------- ------(((...-.))).....-.....................(((((((((((((((((((.....))))))))..))))...))).))))(((.........)))....-------- ( -34.20, z-score = -2.94, R) >droPer1.super_4 5326257 118 - 7162766 GGCUAAUGCCAA-UGCCAAUGC-CAAUGCGAACGGGAAUGCCGGUGGAGACAUCUGACUGCCCAUGCUGGGCGGCCUCAGGUCAAGUCUGCCGCUCGAGGUUUAGGCCUAAGAACUAAGC (((....)))..-.(((...((-(....(((.(((.....)))((((((((((((((((((((.....))))))..))))))...)))).))))))).)))...)))............. ( -43.70, z-score = -0.83, R) >dp4.chr3 11854405 118 - 19779522 GGCUAAUGCCAA-UGCCAAUGC-CAAUGCGAACGGGAAUGCCGGUGGAGACAUCUGACUGCCCAUGCUGGGCGGCCUCAGGUCAAGUCUGCCGCUCGAGGUUUAGGCCUAAGAACUAAGC (((....)))..-.(((...((-(....(((.(((.....)))((((((((((((((((((((.....))))))..))))))...)))).))))))).)))...)))............. ( -43.70, z-score = -0.83, R) >droAna3.scaffold_13266 17194939 96 - 19884421 ------GGCCAA-CGCCAAUCC-GAGCGGCAAUGGCAACGCCAACGGCGACAUCUGACUGCCCAUACUGGGCAGCCUGAGGUGAAGUCUGCCAGCCAGGGAUUA---------------- ------(((...-.)))(((((-..((((((.((((...))))..(((..(((((.(((((((.....))))))..).)))))..))))))).))...))))).---------------- ( -40.20, z-score = -2.05, R) >droEre2.scaffold_4845 4729476 104 - 22589142 ------GGCCAA-UGCCAAUCC-AAAUAGAAAUUCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUAAGGUGAAGUCUGCCGGCGGAGGACUAACGCAACU-------- ------(((...-.)))..(((-.....(.....).....((..(((((((((((((((((((.....))))))))..))))...))).))))..)).)))...........-------- ( -35.10, z-score = -2.33, R) >droYak2.chr2R 12123253 104 - 21139217 ------GGCCAA-UGCCAAUCC-AAAUAGAAACCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUCUGGGCAGUCUAAGGUGAAGUCUGCCGACGGAGGACUAAAGCAACU-------- ------(((...-.)))..(((-.....(.....).....((..(((((((((((((((((((.....))))))))..))))...))).))))..)).)))...........-------- ( -34.00, z-score = -2.52, R) >droSec1.super_1 5486489 106 + 14215200 ------GGCCAAAUGCCCAUCCGAAAUAGAAUUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAGGACUAACGCAACU-------- ------(((.....)))..((((((......)))......((..(((((((((((((((((((.....))))))))..))))...))).))))..)).)))...........-------- ( -35.10, z-score = -2.07, R) >droSim1.chr2R 6473816 104 + 19596830 ------GGCCAA-UGCCAAUCC-GAAUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAGGACUAACGCAACU-------- ------(((...-.))).....-.........(((...((((...((((((((((((((((((.....))))))))..))))...))).)))))))..)))...........-------- ( -35.50, z-score = -2.40, R) >droWil1.scaffold_180700 882779 95 - 6630534 -------------GCCAAAUGC-CAAUGAGAAUGGAGGU---GAGAGAGAUAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUGAAGCCUGGCCACCAGUGAGUAGCCAAACU-------- -------------......(((-((.......(((.(((---.((.(...(((((((((((((.....))))))))..)))))...))).))).))).)).)))........-------- ( -30.10, z-score = -1.60, R) >droVir3.scaffold_12875 12043044 101 - 20611582 -------GGUCAAUGCCAAUUC-AAAUGGCAAUGGCAAC---ACUGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAGAUGAAGCCUGGCCGCCAAUGAGUAGCCAAACU-------- -------.((((.(((((....-...))))).))))...---...(((..(((((((((((((.....))))))..)))))))..)))((((..(......)..))))....-------- ( -43.30, z-score = -4.13, R) >droMoj3.scaffold_6496 18641525 92 + 26866924 -------------AGUCAAUGC-CAAUUCGAACG---GU---AAUGGUGAUAUCUGACUGCCCAUCCUGGGCAGCCUUAGAUAGAGCCUGGCUGCCGGGGAGUUGCCAAAUU-------- -------------......((.-((((((...((---((---(..(((..(((((((((((((.....))))))..)))))))..)))....)))))..)))))).))....-------- ( -36.30, z-score = -2.57, R) >droGri2.scaffold_15245 16349429 101 - 18325388 -------GAUGAACGCCAAUGC-GAAUGGCAAUGGCAAC---ACUGGCGACAUCUGACUGCCCAUUUUAGGCAGCCUUAGAUGAAGCCUGGCCGCCAGGGAGUAGCCAAACU-------- -------.......((((.(((-.....))).))))...---...(((..((((((((((((.......)))))..)))))))..)))((((..(......)..))))....-------- ( -38.10, z-score = -2.77, R) >consensus ______GGCCAA_UGCCAAUCC_AAAUGGAAAUGGAAAUGCCAACGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAGGUGAAGUCUGCCGGCCGAGGACUAGCCCAACU________ ................................((...........(....)((((((((((((.....))))))..)))))).............))....................... (-14.65 = -13.38 + -1.26)


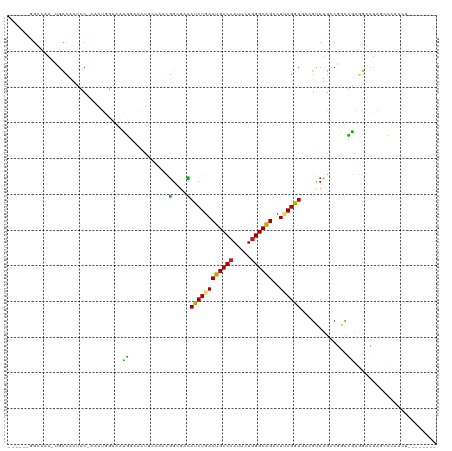
| Location | 7,936,621 – 7,936,725 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.62154 |
| G+C content | 0.53506 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7936621 104 - 21146708 --------AGUUGCGUUAGUUUUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAUUU-GGAUUGGCA-UUGGCC------ --------....((((((((((.((...)).)))))........((((((((.....)))))))).))))).((((.((.((((..(((....))).-.)).)).))-.)))).------ ( -34.60, z-score = -1.68, R) >droPer1.super_4 5326257 118 + 7162766 GCUUAGUUCUUAGGCCUAAACCUCGAGCGGCAGACUUGACCUGAGGCCGCCCAGCAUGGGCAGUCAGAUGUCUCCACCGGCAUUCCCGUUCGCAUUG-GCAUUGGCA-UUGGCAUUAGCC (((.(((.((...(((....((.((((((((((.......))).(((.((((.....)))).))).((((((......)))))).)))))))....)-)....))).-..)).)))))). ( -38.20, z-score = 0.20, R) >dp4.chr3 11854405 118 + 19779522 GCUUAGUUCUUAGGCCUAAACCUCGAGCGGCAGACUUGACCUGAGGCCGCCCAGCAUGGGCAGUCAGAUGUCUCCACCGGCAUUCCCGUUCGCAUUG-GCAUUGGCA-UUGGCAUUAGCC (((.(((.((...(((....((.((((((((((.......))).(((.((((.....)))).))).((((((......)))))).)))))))....)-)....))).-..)).)))))). ( -38.20, z-score = 0.20, R) >droAna3.scaffold_13266 17194939 96 + 19884421 ----------------UAAUCCCUGGCUGGCAGACUUCACCUCAGGCUGCCCAGUAUGGGCAGUCAGAUGUCGCCGUUGGCGUUGCCAUUGCCGCUC-GGAUUGGCG-UUGGCC------ ----------------.(((((.((((((((((.....((.((.((((((((.....)))))))).)).))((((...))))))))))..))))...-)))))(((.-...)))------ ( -40.40, z-score = -1.27, R) >droEre2.scaffold_4845 4729476 104 + 22589142 --------AGUUGCGUUAGUCCUCCGCCGGCAGACUUCACCUUAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGAAUUUCUAUUU-GGAUUGGCA-UUGGCC------ --------......((((((((.(((.((((.(((.((......((((((((.....)))))))).)).))))))).))).....((((....))))-)))))))).-......------ ( -37.20, z-score = -2.38, R) >droYak2.chr2R 12123253 104 + 21139217 --------AGUUGCUUUAGUCCUCCGUCGGCAGACUUCACCUUAGACUGCCCAGAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGGUUUCUAUUU-GGAUUGGCA-UUGGCC------ --------...((((..(((((...(((....)))...((((..((((((((.....))))))))((((((((....))))))))))))........-)))))))))-......------ ( -36.30, z-score = -1.62, R) >droSec1.super_1 5486489 106 - 14215200 --------AGUUGCGUUAGUCCUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAAUUCUAUUUCGGAUGGGCAUUUGGCC------ --------..........(((((((((((((.(((.....((..((((((((.....))))))))))..)))...)))))).....(((......)))))).))))........------ ( -38.20, z-score = -1.97, R) >droSim1.chr2R 6473816 104 - 19596830 --------AGUUGCGUUAGUCCUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAUUC-GGAUUGGCA-UUGGCC------ --------......((((((((.(((.((((.(((.....((..((((((((.....))))))))))..))))))).))).....((((....))))-)))))))).-......------ ( -38.60, z-score = -2.49, R) >droWil1.scaffold_180700 882779 95 + 6630534 --------AGUUUGGCUACUCACUGGUGGCCAGGCUUCACCUAAGACUGCCCAAAAUGGGCAGUCAGAUAUCUCUCUC---ACCUCCAUUCUCAUUG-GCAUUUGGC------------- --------((((((((((((....))))))))))))...((...((((((((.....)))))))).............---....(((.......))-).....)).------------- ( -34.70, z-score = -3.32, R) >droVir3.scaffold_12875 12043044 101 + 20611582 --------AGUUUGGCUACUCAUUGGCGGCCAGGCUUCAUCUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCAGU---GUUGCCAUUGCCAUUU-GAAUUGGCAUUGACC------- --------....((((....((((((((((........((((..((((((((.....)))))))))))))))))))))---)..)))).(((((...-....)))))......------- ( -44.70, z-score = -3.50, R) >droMoj3.scaffold_6496 18641525 92 - 26866924 --------AAUUUGGCAACUCCCCGGCAGCCAGGCUCUAUCUAAGGCUGCCCAGGAUGGGCAGUCAGAUAUCACCAUU---AC---CGUUCGAAUUG-GCAUUGACU------------- --------....((.(((.((..(((......((...(((((..((((((((.....)))))))))))))...))...---.)---))...)).)))-.))......------------- ( -26.70, z-score = -0.28, R) >droGri2.scaffold_15245 16349429 101 + 18325388 --------AGUUUGGCUACUCCCUGGCGGCCAGGCUUCAUCUAAGGCUGCCUAAAAUGGGCAGUCAGAUGUCGCCAGU---GUUGCCAUUGCCAUUC-GCAUUGGCGUUCAUC------- --------....((((..((....))..))))(((..(((((..((((((((.....)))))))))))))..))).((---(.(((((.(((.....-))).)))))..))).------- ( -43.10, z-score = -3.44, R) >consensus ________AGUUGGGCUAGUCCUCGGCCGGCAGACUUCACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUCCAUUUCCAUUU_GGAUUGGCA_UUGGCC______ ............................................((((((((.....)))))))).......((((..........................)))).............. (-16.39 = -16.53 + 0.14)
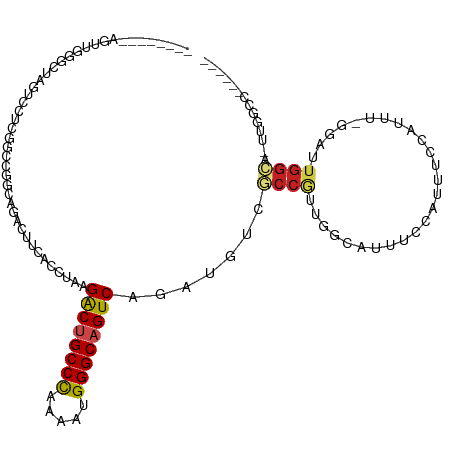

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:42 2011