| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,603,778 – 2,603,876 |
| Length | 98 |
| Max. P | 0.929615 |

| Location | 2,603,778 – 2,603,876 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.06 |
| Shannon entropy | 0.58726 |
| G+C content | 0.43038 |
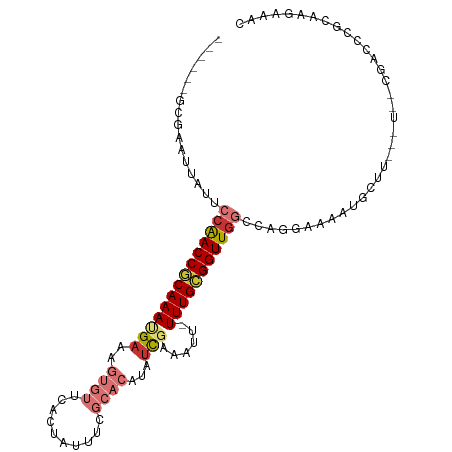
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -12.14 |
| Energy contribution | -13.18 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
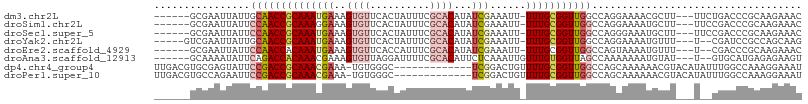
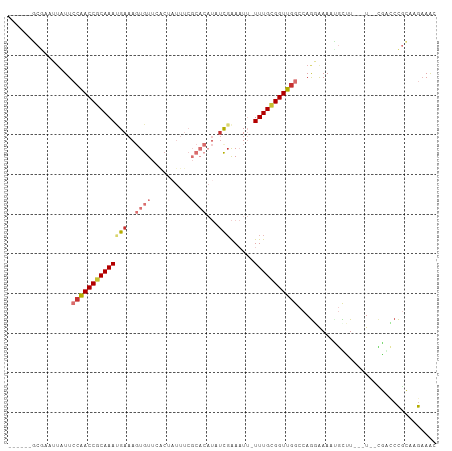
>dm3.chr2L 2603778 98 - 23011544 ------GCGAAUUAUUGCAACCGCAAAUGAAAGUGUUCACUAUUUCGCACAUAUCGAAAUU-UUUGCGGUUGGCCAGGAAAACGCUU---UUCUGACCCGCAAGAAAC ------(((........((((((((((((((....))))..((((((.......)))))).-))))))))))..(((((((....))---)))))...)))....... ( -27.30, z-score = -2.36, R) >droSim1.chr2L 2562269 98 - 22036055 ------GCGAAUUAUUCCAACCGCAAAGGAAAGUGUUCACUAUUUCGCACAUAUCGAAAUU-UUUGCGGUUGGCCAGGAAAAUGCUU---UUCCGACCCGCAAGAAAC ------(((.......(((((((((((((..(((....))).(((((.......)))))))-)))))))))))...(((((.....)---))))....)))....... ( -29.50, z-score = -3.14, R) >droSec1.super_5 772125 98 - 5866729 ------GCGAAUUAUUCCAACCGCAAAUGAAAGUGUUCACUAUUUCGCACAUAUCGAAAUU-UUUGCGGUUGGCCAGGGAAAUGCUU---UUCCGACCCGCAAGAAAC ------(((.......(((((((((((((((....))))..((((((.......)))))).-)))))))))))....(((((....)---))))....)))....... ( -29.20, z-score = -2.92, R) >droYak2.chr2L 2592409 97 - 22324452 -----GUCGAAUUAUUGCAACCGCAAAUGAAAGUGUUCACUAUUUCGCACAUAUCGAAAUU-UUUGCGGUUGGCCAGGAAAAUGUUU---U--CGAUCCGCCAGCAAG -----.........(((((((((((((((((....))))..((((((.......)))))).-)))))))))(((..(((........---.--...)))))).)))). ( -24.00, z-score = -1.18, R) >droEre2.scaffold_4929 2648407 96 - 26641161 ------GCGAAUUAUUCCAACCACAAAUGAAAGUGUUCACCAUUUCGCACAUAUCGAAAUU-UUUGCGGUUGGCCAGUAAAAUGUUU---U--CGACCCGCAAGAAAC ------(((((..........(((........)))........)))))...........((-(((((((((((.((......))...---)--))).))))))))).. ( -19.37, z-score = -0.94, R) >droAna3.scaffold_12913 418886 97 + 441482 ------GCAAAAUAUUCAGACCACAAACGAAAGUGUUAGGAUUUUCGCACAUUCUCAAAUUGUUUGUGGUUAGCCAAAAAAAUGUAU---U--GUGCAUGAGAGAAGU ------........((((((((((((((((...((..(((((........)))))))..)))))))))))).(((((.........)---)--).)).))))...... ( -20.50, z-score = -1.16, R) >dp4.chr4_group4 1912327 94 - 6586962 UUGACGUGCGAGUAUUCCGACCGCAAACGAAA-UGUGGGC-------------UCGGACUGUUUUGCGGUUGGCCAGCAAAAAACGUACAUAUUUGGCCAAAGGAAAU ......((((((...((((((((((.......-)))))..-------------)))))....)))))).((((((((................))))))))....... ( -25.49, z-score = -0.30, R) >droPer1.super_10 919479 94 - 3432795 UUGACGUGCCAGAAUUCCGACCGCAAACGAAA-UGUGGGC-------------UCGGACUGUUUUGCGGUUGGCCAGCAAAAAACGUACAUAUUUGGCCAAAGGAAAU .....(.((((((..((((((((((.......-)))))..-------------)))))...((((((((....)).))))))..........)))))))......... ( -26.50, z-score = -0.73, R) >consensus ______GCGAAUUAUUCCAACCGCAAAUGAAAGUGUUCACUAUUUCGCACAUAUCGAAAUU_UUUGCGGUUGGCCAGGAAAAUGCUU___U__CGACCCGCAAGAAAC ................((((((((((((((..((((..........))))...)))......)))))))))))................................... (-12.14 = -13.18 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:49 2011