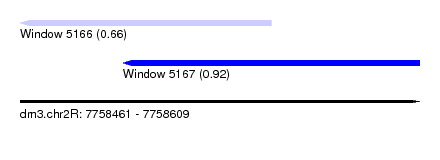
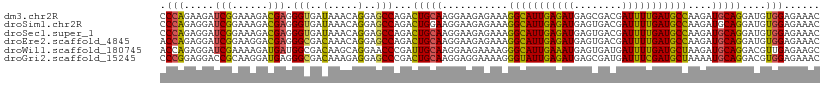
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,758,461 – 7,758,609 |
| Length | 148 |
| Max. P | 0.915100 |

| Location | 7,758,461 – 7,758,554 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.27 |
| Shannon entropy | 0.39458 |
| G+C content | 0.50479 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.16 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
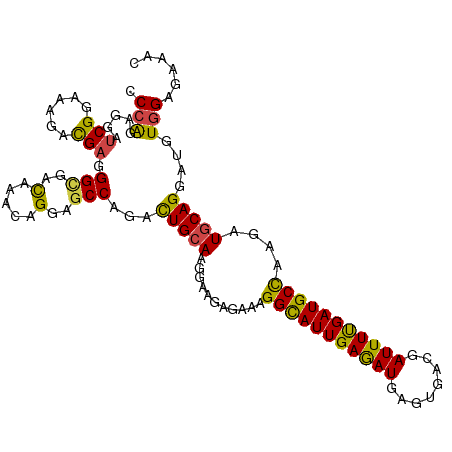
>dm3.chr2R 7758461 93 - 21146708 AAAGGCAUUGAGAUGAGCGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAACC------CGAGGAGGAUGACAGCGGUGAGUCCGAAGAAGAGGAG ...(((((..((((........))))..)))))........((((((......))(------((..(.......)..)))...))))............ ( -19.20, z-score = -1.19, R) >droSim1.chr2R 6295537 93 - 19596830 AAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAACC------CGAGGAGGAUGACAGCGGUGAGUCCGAAGAAGAGGAG ...(((((..((((........))))..)))))........((((((......))(------((..(.......)..)))...))))............ ( -19.20, z-score = -1.47, R) >droSec1.super_1 5309942 93 - 14215200 AAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAACC------CGAGGAGGAUGACAGCGGUGAGUCCGAAGAAGAGGAG ...(((((..((((........))))..)))))........((((((......))(------((..(.......)..)))...))))............ ( -19.20, z-score = -1.47, R) >droEre2.scaffold_4845 4551982 93 + 22589142 AAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAACC------CGAGGAGGAUGACAGCGGUGAGUCCGAGGAAGAGGAG ...(((((..((((........))))..)))))......................(------(..(((...(.((....)).).)))..))........ ( -19.30, z-score = -1.25, R) >droYak2.chr2R 11945599 93 + 21139217 AAAGGCAUUGAGAUGAGCGACGAUUUUGAUGCCAAGAUGCAAGAUGUGGAGAAACC------UGAGGAGGAUGACAGCGGUGAGUCCGAGGAUGAGGAG ...(((((..((((........))))..))))).....................((------(.(...((((.((....))..)))).....).))).. ( -21.20, z-score = -1.86, R) >droVir3.scaffold_12823 1781875 99 + 2474545 AAGGGCAUCGAAAUGAGCGACGACUUCGAUGCUAAAAUGCAGGACGUGGAGAAACCAGAGAACGAGGAGGAUGACAGUGGCGAAUCUGAAAAUGAAGAU ................((.((..(((((.(((......))).....(((.....))).....))))).(.....).)).))..((((........)))) ( -15.90, z-score = 0.51, R) >droMoj3.scaffold_6496 3267029 99 - 26866924 AAGGGCAUCGAAAUGAGCGAUGAUUUCGAUGCCAAAAUGCAGGACGUGGAAAAGCCAGAGAACGAGGAGGAUGACAGUGGCGAAUCCGAGGAUGAGGAG ...(((((((((((.(....).)))))))))))......((...(.((((...((((....................))))...)))).)..))..... ( -26.35, z-score = -2.48, R) >anoGam1.chr2L 45687528 96 - 48795086 AAGGGAAUCGAUAUGAGUGAAGAUUUCGAUUCAAAGCUGCAGGAUAUGGAACGGCCGGAGGGUUCGGACUCGGAGGAAGAAAAGGACGAAGACGAG--- ....(((((((.((........)).)))))))...((((............))))....(..((((..((............))..))))..)...--- ( -16.70, z-score = 1.12, R) >consensus AAAGGCAUUGAGAUGAGCGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAACC______CGAGGAGGAUGACAGCGGUGAGUCCGAAGAAGAGGAG ...(((((((((((........)))))))))))...................................((((...........))))............ (-15.16 = -14.16 + -1.00)
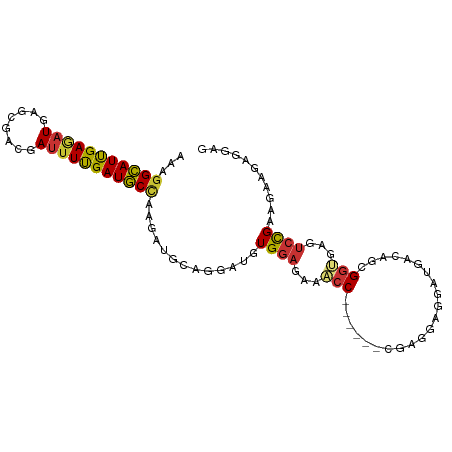
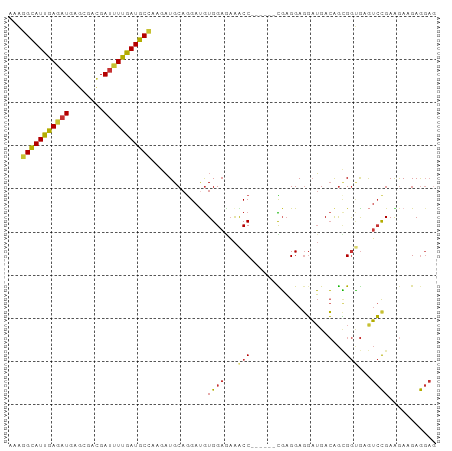
| Location | 7,758,499 – 7,758,609 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Shannon entropy | 0.20759 |
| G+C content | 0.49545 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -18.75 |
| Energy contribution | -17.86 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7758499 110 - 21146708 CCCAGAAGAUCGGAAAGACGAGGGUGAUAAACAGGAGCCAGACUGCAAGGAAGAGAAAGGCAUUGAGAUGAGCGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAAC .(((.....(((......))).................((..(((((...........(((((..((((........))))..)))))....)))))..)))))...... ( -23.16, z-score = -2.04, R) >droSim1.chr2R 6295575 110 - 19596830 CCCAGAGGAUCGGAAAGACGAGGGUGAUAAACAGGAGCCAGACUGGAAGGAAGAGAAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAAC .((((....(((......))).(((..(.....)..)))...))))............(((((..((((........))))..)))))...................... ( -23.00, z-score = -2.16, R) >droSec1.super_1 5309980 110 - 14215200 CCCAGAGGAUCGGAAAGACGAGGGUGAUAAACAGGAGCCAGACUGCAAGGAAGAGAAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAAC .(((.....(((......))).................((..(((((...........(((((..((((........))))..)))))....)))))..)))))...... ( -23.16, z-score = -1.93, R) >droEre2.scaffold_4845 4552020 110 + 22589142 ACCAGAGGAUCGGAAGGACGAGGGCGACAAACAGGAGCCAGACUGCAAGGAAGAGAAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAAC .(((.(...(((......))).(((..(.....)..)))...(((((...........(((((..((((........))))..)))))....)))))..).)))...... ( -27.16, z-score = -3.14, R) >droWil1.scaffold_180745 105303 110 + 2843958 ACCAGAGGAUCGAAAAGAUGAUGGCGACAAGCAGGAACCCGAUUGCAAGGAAGAAAAGGGCAUUGAAAUGAGUGAUGAUUUUGAUGCUAAGAUGCAGGACGUUGAGAAGC .(((....(((.....)))..)))((((..((((........))))............(((((..((((.(....).))))..)))))............))))...... ( -22.50, z-score = -1.53, R) >droGri2.scaffold_15245 8945700 110 - 18325388 CCCGGAGGACCGCAAGGAUGAGGGCGACAAAGAGGAGCCCGACUGCAAGGAGGAAAAGGGUAUUGAGAUGAGCGAUGAUUUCGAUGCUAAAAUGCAGGACGUGGAGAAAC ((((.....((....))....((((..(.....)..))))..(((((...........(((((((((((.(....).)))))))))))....)))))..)).))...... ( -33.06, z-score = -3.64, R) >consensus CCCAGAGGAUCGGAAAGACGAGGGCGACAAACAGGAGCCAGACUGCAAGGAAGAGAAAGGCAUUGAGAUGAGUGACGAUUUUGAUGCCAAGAUGCAGGAUGUGGAGAAAC .(((.....(((......))).(((..(.....)..)))...(((((...........(((((((((((........)))))))))))....)))))....)))...... (-18.75 = -17.86 + -0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:24 2011