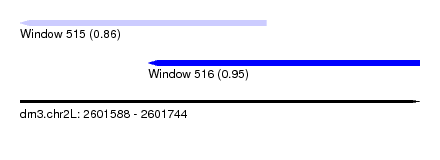
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,601,588 – 2,601,744 |
| Length | 156 |
| Max. P | 0.951712 |

| Location | 2,601,588 – 2,601,684 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.42 |
| Shannon entropy | 0.66450 |
| G+C content | 0.55189 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -11.67 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
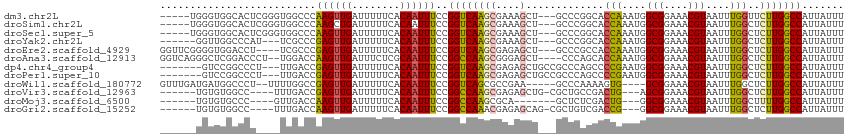
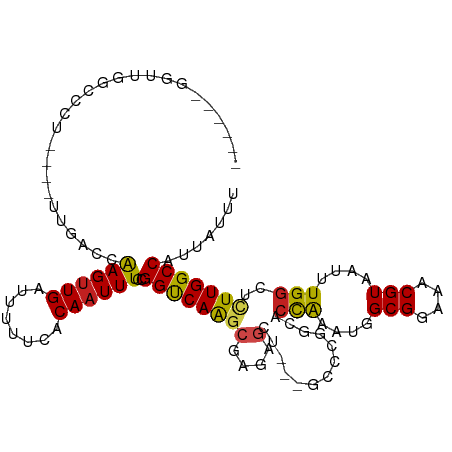
>dm3.chr2L 2601588 96 - 23011544 CCGGCACCAAAUG-GCGGAAACGUAAUUUGGUUCUUGGCCAUUA-UUUUCGGUUCUGGCCGCAAA-GCCAAAGC--GAACUCAUCGUCCUUCGAUGGUCGG-------- ..((((((((((.-(((....))).)))))))....(((((..(-(.....))..))))).....-)))....(--((.(.(((((.....))))))))).-------- ( -36.10, z-score = -3.03, R) >droSim1.chr2L 2560066 96 - 22036055 CCGGCACCAAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGUUCUGGCCGCAAA-GCCAAAGC--GAACUCAUCGUCCUUCGAUGGGCGC-------- ..(((.((((((.-(((....))).)))))).....(((((..(-(.....))..))))).....-)))...((--(..(((((((.....))))))))))-------- ( -37.50, z-score = -2.80, R) >droSec1.super_5 769941 96 - 5866729 CCGGCACCAAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGUUCUGGCCGCAAA-GCCAAAGC--GAACUCAUCGUCCUUCGAUGGGCGG-------- (((((........-(((....)))..(((((((..((((((..(-(.....))..))))))...)-))))))))--...(((((((.....))))))))))-------- ( -36.60, z-score = -2.57, R) >droYak2.chr2L 2590038 97 - 22324452 CCGGCACCAAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUUCGGUUCUGGCCGCAAU-GCCAAAGC--GUACUCAUCGUCCUUCGAUGGGCGG-------- ((((((((((((.-(((....))).)))))).....(((((..((......))..)))))....)-))).....--...(((((((.....))))))).))-------- ( -36.60, z-score = -2.61, R) >droEre2.scaffold_4929 2646104 96 - 26641161 CCGCCACCAAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGUUCUGGCCGCAAA-GCCAAAGC--GAACUCAUAGUCCUUCGAUGGGCGG-------- ((((((.....))-))))...(((..(((((((..((((((..(-(.....))..))))))...)-))))))))--)........((((......))))..-------- ( -35.90, z-score = -2.58, R) >droAna3.scaffold_12913 415805 103 + 441482 CCAGCACCAAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGUUCGGACCACAAACACCCCAGCUACAGCGGAUUG-GUUUCGAGGGGCGGUCGGU--- ((((..((((((.-(((....))).))))))..)).((((....-.....(((....))).......((((.((....))(((...-..)))..)))).)))))).--- ( -32.20, z-score = -0.44, R) >dp4.chr4_group4 1908716 86 - 6586962 CCAGCCCCGAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUUGGCUCUGACCGCAG--ACCCAAGCGGCCAGGGGAGGGCAG------------------- ...(((((((((.-(((....))).))))).(((((((((....-..(((((.((((....)))--).))))).))))))))).))))..------------------- ( -40.00, z-score = -3.14, R) >droPer1.super_10 915960 86 - 3432795 CCAGCCCCGAAUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUUGGCUCUGACCGCAG--ACCCAAGCGGCCAGGGGAGGGCAG------------------- ...(((((((((.-(((....))).))))).(((((((((....-..(((((.((((....)))--).))))).))))))))).))))..------------------- ( -40.00, z-score = -3.14, R) >droWil1.scaffold_180772 1253078 79 - 8906247 ----CCAAAAGUG-GCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGCGCUGACCGCAAUGUUCCUCCUCUCCGUCUCCC------------------------ ----......(.(-(((((..((.(((.((((.....))))..)-))..))(((.....))).............)))))).)..------------------------ ( -18.30, z-score = -0.33, R) >droVir3.scaffold_12963 11178406 106 - 20206255 -CGCUGCCGACUGAGCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGCACUGACCGCAAA-GCCAAAGUCCGCUGCAGCUGCCGCUCUACGAGUGGACAGGGGA -.((.((.(((((((((....)))....((((.....))))...-...))(((..((....))..-)))..)))).)).))..(((((((((...)))))).))).... ( -37.00, z-score = -0.50, R) >droMoj3.scaffold_6500 19911772 102 - 32352404 --GCUCUCGACUGGGCGGAAACGUAAUUUGGCUUUUGGCCAUUA-UUUUCGGCACUGACCACAAA-GCCAAAGUCCGCUGCAGCUGC--CUGCAGGAGCGAAGGGAGC- --((((((((((..(((....)))...(((((((((((.((...-..........)).))).)))-)))))))))((((.(.((...--..)).).))))..))))))- ( -38.62, z-score = -1.57, R) >droGri2.scaffold_15252 5311389 99 - 17193109 -CGCUGUCGACCGGGCGGAAACGUAAUUUGGCUCUUGGCCAUUA-UUUUCGGCACUGACCACAAA-GCCAAAGUCCACUGCAGCUGC----GUAGCUGCGUAGGGU--- -........((((((((....)....(((((((.((((((....-.....)))........))))-))))))))))...((((((..----..))))))....)))--- ( -30.80, z-score = 0.29, R) >consensus CCGGCACCAAAUG_GCGGAAACGUAAUUUGGCUCUUGGCCAUUA_UUUUCGGCUCUGACCGCAAA_GCCAAAGC__GCACUCAUCGUCCUUCGAUGGGCGG________ ..............(((....)))....((((.....)))).........((......))................................................. (-11.67 = -11.59 + -0.08)
| Location | 2,601,638 – 2,601,744 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Shannon entropy | 0.44736 |
| G+C content | 0.50676 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.59 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2601638 106 - 23011544 -----UGGGUGGCACUCGGGUGGCCCAAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAAAGCU---GCCCGGCACCAAAUGGCGGAAACGUAAUUUGGUUCUUGGCCAUUAUUU -----.(((..((..(((..(((((.((((((........))))))..)))))..)))..)).---.)))((((((((((.(((....))).))))))).....)))....... ( -43.70, z-score = -3.41, R) >droSim1.chr2L 2560116 106 - 22036055 -----UGGGUGGCACUCGGGUGGCCCAAGCUGAUUUUUCACAAUUUCCGGUCAAGCGAAAGCU---GCCCGGCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU -----.(((..((..(((..(((((.....(((....)))........)))))..)))..)).---.)))(((.((((((.(((....))).))))))......)))....... ( -39.32, z-score = -1.74, R) >droSec1.super_5 769991 106 - 5866729 -----UGGGUGGCACUCGGGUGGCCCAAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAAAGCU---GCCCGGCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU -----.(((..((..(((..(((((.((((((........))))))..)))))..)))..)).---.)))(((.((((((.(((....))).))))))......)))....... ( -42.40, z-score = -2.83, R) >droYak2.chr2L 2590089 102 - 22324452 ------GGUUGGCCCAU---UCGCCCGAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAAAGCU---GCCCGGCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ------...(((((...---..(((.((((((........))))))..((...(((....)))---..))))).((((((.(((....))).)))))).....)))))...... ( -36.20, z-score = -2.28, R) >droEre2.scaffold_4929 2646154 107 - 26641161 GGUUCGGGGUGGACCU----UCGCCCGAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAGAGCU---GCCCGCCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ......(((..(.(.(----((((((((((((........)))))..)))....))))).)).---.)))((((((((((.(((....))).))))))....))))........ ( -36.40, z-score = -1.21, R) >droAna3.scaffold_12913 415862 108 + 441482 GGUCAGGGCUCGGACCCU--UGGACCAAGUUGAUUUCUCGCAAUUUCCGGCCAAGCGGGAGCU----CCCAGCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ((((((((((.(((((((--(((.((((((((........))))))..))))))).))....)----)).))).((((((.(((....))).))))))..)))))))....... ( -42.90, z-score = -2.61, R) >dp4.chr4_group4 1908756 104 - 6586962 -------GUCCGGCCCU---UUGACCGAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAGAGCUGCCGCCCAGCCCCGAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU -------....(((...---.(((((((((((........)))))..))))))(((....))))))(((.((..((((((.(((....))).))))))..)).)))........ ( -38.40, z-score = -3.11, R) >droPer1.super_10 916000 104 - 3432795 -------GUCCGGCCCU---UUGACCGAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAGAGCUGCCGCCCAGCCCCGAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU -------....(((...---.(((((((((((........)))))..))))))(((....))))))(((.((..((((((.(((....))).))))))..)).)))........ ( -38.40, z-score = -3.11, R) >droWil1.scaffold_180772 1253115 103 - 8906247 GUUUGAUGAUGGCCCU--UUUUGGCCGAGUUGAUUUUUCACAAUUUCCGGUCAGCGCCGAA-----GCCCAAAAGUG----GCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ....((((((((((..--..((((((((((((........)))))..))))))).((((((-----((((....).)----))(....)....))))))....)))))))))). ( -37.30, z-score = -3.02, R) >droVir3.scaffold_12963 11178466 100 - 20206255 ------UGUGUGGCC----UUUGACCGAGUUGAUUUUUCACAAUUUCCGGCCAAGCGAGAGCUG-CGCUGCCGACUG---AGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ------...((((((----...(((((((((................((((..(((....))).-....))))....---.(((....)))))))))).))..))))))..... ( -34.40, z-score = -2.18, R) >droMoj3.scaffold_6500 19911829 94 - 32352404 ------UGUGUGCCC----GUUGACCAAGUUGAUUUUUCACAAUUUCCGGCCAAGCGCA-------GCUCUCGACUG---GGCGGAAACGUAAUUUGGCUUUUGGCCAUUAUUU ------((((.....----((..((...))..))....))))......(((((((.((.-------((((......)---)))(....)........)).)))))))....... ( -26.90, z-score = -0.72, R) >droGri2.scaffold_15252 5311442 100 - 17193109 ------UGUGUGGCC----UUUGACCAAGUUGAUUUUUCACAAUUUCCGGCCAAACGAGAGCAG-CGCUGUCGACCG---GGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ------...((((((----...(((((((((((....))....((((((.((...(((.(((..-.))).)))...)---).))))))...))))))).))..))))))..... ( -36.40, z-score = -2.74, R) >consensus ______GGUUGGCCCU____UUGACCAAGUUGAUUUUUCACAAUUUCCGGUCAAGCGAGAGCU___GCCCGGCACCAAAUGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUU ..........................((((((........))))))..((((((((....).............(((....(((....)))....)))..)))))))....... (-18.14 = -17.59 + -0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:49 2011