| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,730,772 – 7,730,878 |
| Length | 106 |
| Max. P | 0.798988 |

| Location | 7,730,772 – 7,730,878 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 60.63 |
| Shannon entropy | 0.86208 |
| G+C content | 0.40956 |
| Mean single sequence MFE | -16.71 |
| Consensus MFE | -6.55 |
| Energy contribution | -6.39 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.798988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
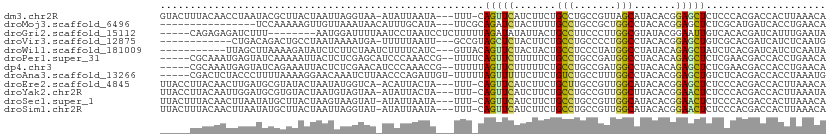
>dm3.chr2R 7730772 106 - 21146708 GUACUUUACAACCUAAUACGCUUACUAAUUAGGUAA-AUAUUAAUA---UUU-CAGUUCAUCUUCUGCCUGCCGUUAGCAUACACGGAGCUCUCCCACGACCACUUAAACA ..........(((((((..........)))))))..-.........---...-..(((........((...((((........)))).))........))).......... ( -11.09, z-score = 0.51, R) >droMoj3.scaffold_6496 2153964 92 + 26866924 ----------------UCCAAAAAGUUGUUAAAUAACAUUUGCAUA---UUCGCAGAUCUACUUUUGCCUGCCGCUGGCCUACACGGAGCUCUCGCAUGAUCACCUGAACA ----------------(((.((((((((((....))))(((((...---...)))))...))))))(((.......)))......)))((....))............... ( -15.10, z-score = 0.50, R) >droGri2.scaffold_15112 375913 98 - 5172618 -----CAGAGAGAUCUUU--------AAUGGAUUUUAAUCCUAAUCCUCUUUUUAGAUAUAUUACUGCCUUCCCUUGGCGUAUACGGAAUUGUCACACGAUCAUUUGAAUA -----((((..((((...--------...((((....))))...((((((....))).....(((.(((.......))))))...)))..........)))).)))).... ( -17.10, z-score = -0.54, R) >droVir3.scaffold_12875 5379006 95 - 20611582 ------------CUGACAGACUGCCUAAUAAAAUGA-UUUUUAAUU---GCCGUAGCUCUACUUCUGCCUGCCCCUGGCCUACACGGAGCUGUCGCACGAUCAUCUCAAUG ------------....................((((-((......(---((..(((((((......(((.......)))......)))))))..))).))))))....... ( -19.00, z-score = -1.06, R) >droWil1.scaffold_181009 3369334 97 + 3585778 -----------UUAGCUUAAAAGAUAUCUCUUCUAAUCUUUUCAUC---GUUACAGUUCUACUACUGCCUCCCUAUGGCCUAUACAGAGCUAUCUCACGAUCAUCUCAAUA -----------.......(((((((..........))))))).(((---((...((((((......(((.......)))......)))))).....))))).......... ( -14.50, z-score = -1.79, R) >droPer1.super_31 162206 104 + 935084 -----CGCAAAUGAGUAUCAAAAAUUACUCUCGAGCAUCCCAAACCG--UUUUCAGUUCUUUUUCUGCCUGCCGAUGGCCUACACAGAGCUCUCGAACGACCACCUGAACA -----.((....(((((........)))))....)).........((--(((..((((((......(((.......)))......))))))...)))))............ ( -20.90, z-score = -1.42, R) >dp4.chr3 16986686 104 - 19779522 -----CGCAAAUGAGUAUCAGAAAUUACUCUCGAACAUCCCAAACCG--UUUUUAGUUCUUUUUCUGCCUGCCGAUGGCCUACACAGAGCUCUCGAACGACCACCUGAACA -----.......(((((........)))))...............((--(((..((((((......(((.......)))......))))))...)))))............ ( -18.30, z-score = -0.69, R) >droAna3.scaffold_13266 6709854 105 + 19884421 -----CGACUCUACCCUUUUAAAAGGAACAAAUCUUAACCCAGAUUGU-UUUUUAGUUUUUCUUCUGUCUGCCUUUGGCCUACACGGAGCUGUCUCACGACCACCUAAAUG -----.(((.............(((((((((.(((......)))))))-))))).......(((((((..(((...)))..))).))))..)))................. ( -16.30, z-score = -0.19, R) >droEre2.scaffold_4845 4525090 106 + 22589142 UUACCUUACAACUUGAUGCGUAUACUAAUAUGGUCA-ACAUUACUA---UUU-CAGUUCAUCUUCUGCUUGCCGUUGGCAUACACGGAGCUCUCCCACGACCACUUAAACA ..............................(((((.-.....(((.---...-.)))....((((((..((((...))))..)).)))).........)))))........ ( -14.60, z-score = 0.21, R) >droYak2.chr2R 11918351 106 + 21139217 UUACCUUACAAUUGGAUGCGUGUACUAAUGUAGUAA-AUAUUACUA---UUU-CAGUUCAUCUUCUGCCUGCCGUUGGCUUACACGGAACUCUCCCACGACCACUUAAAUA ............(((...((((.......((((((.-....)))))---)..-.(((((.......(((.......))).......)))))....)))).)))........ ( -19.04, z-score = -0.88, R) >droSec1.super_1 5282520 106 - 14215200 UUACUUUACAACUUAAUAUGCUUACUAAGUAAGUAU-AUAUUAAUA---UUU-CAGUUCAUCUUCUGCCUGCCGUUGGCAUACACGGAACUCUCCCACGACCACUUAAACA ...............((((((((((...))))))))-)).......---...-.(((((......((((.......))))......))))).................... ( -18.40, z-score = -2.55, R) >droSim1.chr2R 6263867 106 - 19596830 UUACUUUACAACUUAAUAUGCUUACUAAUUAGGUAU-AUAUUAAUA---UUU-CAGUUCAUCUUCUGCCUGCCGUUGGCAUACACGGAACUCUCCCACGACCACUUAAACA ...............(((((((((.....)))))))-)).......---...-.(((((......((((.......))))......))))).................... ( -16.20, z-score = -1.77, R) >consensus _____UUACAAUUUGAUUCAAAUACUAAUUAAGUAA_AUAUUAAUA___UUU_CAGUUCUUCUUCUGCCUGCCGUUGGCCUACACGGAGCUCUCCCACGACCACCUAAACA ......................................................(((((.......(((.......))).......))))).................... ( -6.55 = -6.39 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:22 2011