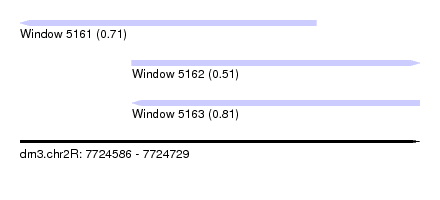
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,724,586 – 7,724,729 |
| Length | 143 |
| Max. P | 0.812913 |

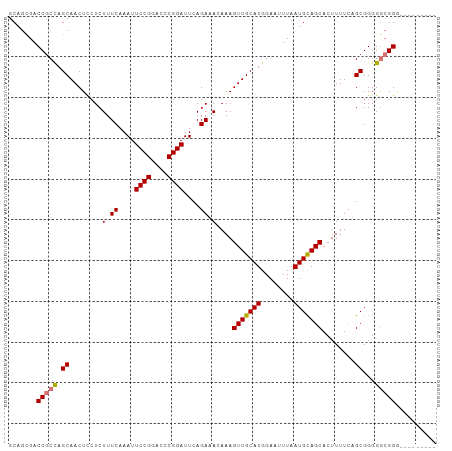
| Location | 7,724,586 – 7,724,692 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Shannon entropy | 0.23282 |
| G+C content | 0.51469 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -27.19 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
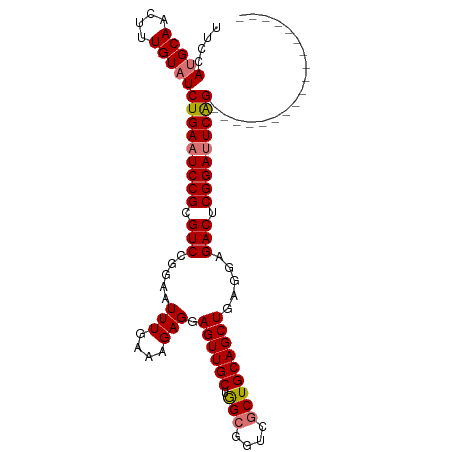
>dm3.chr2R 7724586 106 - 21146708 UUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUAGCGGUCGCUGCAGCUGAGGAGACUCGGAUUCAGAUUCAGAUUCGGAUUCGG ....(((((....)))))(((((((((.(((..(((((((((.....((((((.(((....)))))))))(((....)))...))))))))).))).))))))))) ( -43.70, z-score = -3.58, R) >droSim1.chr2R 6257682 88 - 19596830 UUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCGCUGCAGCUGAGGAGACUCGGAUUCGG------------------ ....(((((....)))))((((((((.....................((((((.(((....)))))))))(((....)))))))))))------------------ ( -29.20, z-score = -0.76, R) >droSec1.super_1 5276430 88 - 14215200 UUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCGCUGCAGCUGAGGAGACUCGGAUUCGG------------------ ....(((((....)))))((((((((.....................((((((.(((....)))))))))(((....)))))))))))------------------ ( -29.20, z-score = -0.76, R) >droEre2.scaffold_4845 4518924 106 + 22589142 UUCCAUGCAACUUUGUUUCUGAAUCCGGGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCACUGCAGCUGAAUAGACUCGGACUCAGAUUCAGAUUCGGAUUCGG ..................(((((((((((((..((((((((.....(((((((((.(((...)))))))......)))))....)))))))).))))))))))))) ( -35.60, z-score = -1.59, R) >consensus UUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCGCUGCAGCUGAGGAGACUCGGAUUCAG__________________ ....(((((....)))))(((((((((.(((.....(((....))).((((((.(((....))))))))).....))).))))))))).................. (-27.19 = -27.50 + 0.31)

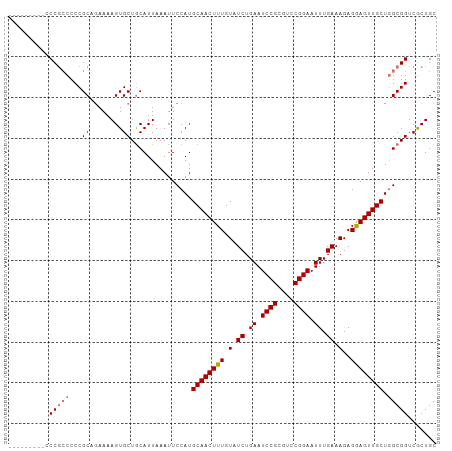
| Location | 7,724,626 – 7,724,729 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Shannon entropy | 0.19451 |
| G+C content | 0.53695 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -25.50 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7724626 103 + 21146708 GCAGCGACCGCUAGCAACUCCUCUUUCAAAUUCCGGACGCGGAUUCAGAUACAAAGUUGCAUGGAAUUUAAUGUAGCACUUUUGAGCGGGGGCGGUAGCGGGG-- .......((((((.(...(((((.((((((.((((....))))............(((((((........)))))))...)))))).))))).).))))))..-- ( -33.50, z-score = -1.64, R) >droSim1.chr2R 6257704 105 + 19596830 GCAGCGACCGCCAGCAACUCCUCUUUCAAAUUCCGGACGCGGAUUCAGAUACAAAGUUGCAUGGAAUUUAAUGCAGCACUUUUCAGCGGGGCGGGGGCGGGGGCG ...((..(((((..(..((((.((.((....((((....))))....))......(((((((........))))))).......)).))))..).)))))..)). ( -36.60, z-score = -1.65, R) >droYak2.chr2R 11912108 96 - 21139217 GCAGCGACCGCCAGCAACUUCUCUUUCAAAUUCCGGGCCCGGAUUCAGAAACAAAGUUGCAUGGAAUUUAAUGCAGCACUUUUCUGCGGGGGCGGG--------- .......(((((.(((((((...((((....((((....))))....))))..)))))))...........(((((.......)))))..))))).--------- ( -32.60, z-score = -1.74, R) >droEre2.scaffold_4845 4518964 96 - 22589142 GCAGUGACCGCCAGCAACUCCUCUUUCAAAUUCCGGACCCGGAUUCAGAAACAAAGUUGCAUGGAAUUUAAUGCAGCACUUUUCUGCGGGGGCGGG--------- .......(((((.(((.......((((....((((....))))....))))....(((((((........))))))).......)))...))))).--------- ( -31.90, z-score = -1.84, R) >consensus GCAGCGACCGCCAGCAACUCCUCUUUCAAAUUCCGGACCCGGAUUCAGAAACAAAGUUGCAUGGAAUUUAAUGCAGCACUUUUCAGCGGGGGCGGG_________ .......(((((.((........((((....((((....))))....))))....(((((((........)))))))........))...))))).......... (-25.50 = -26.12 + 0.62)

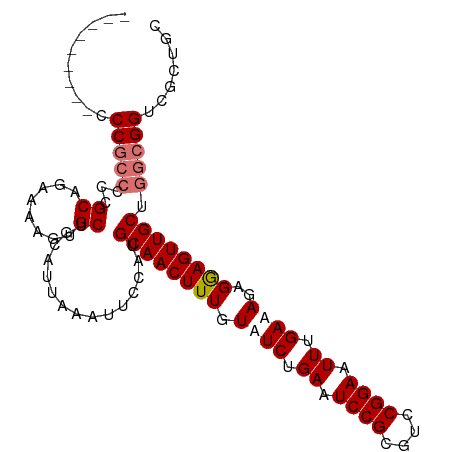
| Location | 7,724,626 – 7,724,729 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Shannon entropy | 0.19451 |
| G+C content | 0.53695 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.92 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
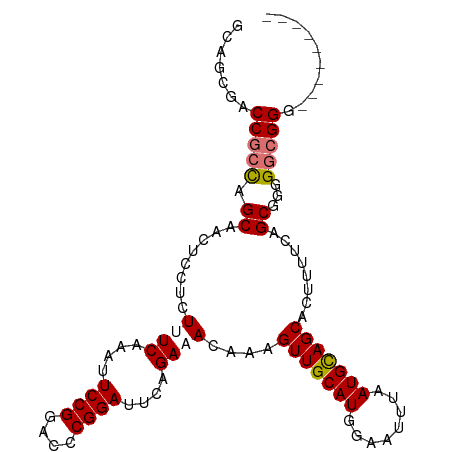
>dm3.chr2R 7724626 103 - 21146708 --CCCCGCUACCGCCCCCGCUCAAAAGUGCUACAUUAAAUUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUAGCGGUCGCUGC --....((.(((((...((((....))))................((((((((...((.((.((((....)))).)).))....))))))))..))))).))... ( -27.60, z-score = -1.41, R) >droSim1.chr2R 6257704 105 - 19596830 CGCCCCCGCCCCCGCCCCGCUGAAAAGUGCUGCAUUAAAUUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCGCUGC .((..(((((...((..((((....))))..))............((((((((...((.((.((((....)))).)).))....)))))))).)))))..))... ( -35.50, z-score = -2.53, R) >droYak2.chr2R 11912108 96 + 21139217 ---------CCCGCCCCCGCAGAAAAGUGCUGCAUUAAAUUCCAUGCAACUUUGUUUCUGAAUCCGGGCCCGGAAUUUGAAAGAGAAGUUGCUGGCGGUCGCUGC ---------.(((((...((((.......))))............((((((((.((((.((.((((....)))).)).))))..)))))))).)))))....... ( -34.40, z-score = -2.59, R) >droEre2.scaffold_4845 4518964 96 + 22589142 ---------CCCGCCCCCGCAGAAAAGUGCUGCAUUAAAUUCCAUGCAACUUUGUUUCUGAAUCCGGGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCACUGC ---------.(((((...((((.......))))............((((((((.((((.((.((((....)))).)).))))..)))))))).)))))....... ( -34.00, z-score = -2.42, R) >consensus _________CCCGCCCCCGCAGAAAAGUGCUGCAUUAAAUUCCAUGCAACUUUGUAUCUGAAUCCGCGUCCGGAAUUUGAAAGAGGAGUUGCUGGCGGUCGCUGC ..........(((((...((........))...............((((((((.((((.((.((((....)))).)).))))..)))))))).)))))....... (-23.86 = -24.92 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:20 2011