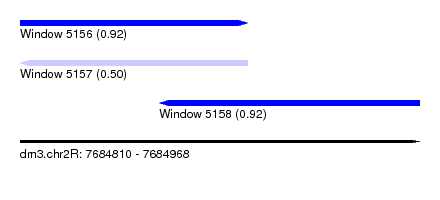
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,684,810 – 7,684,968 |
| Length | 158 |
| Max. P | 0.923150 |

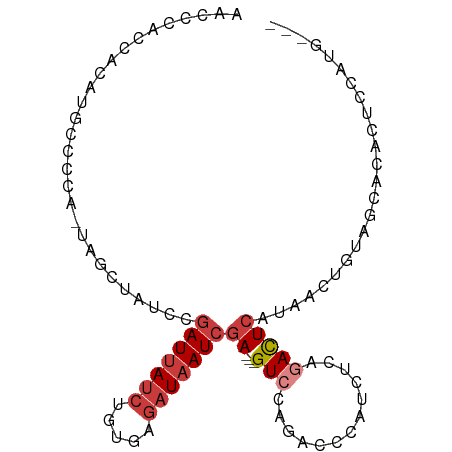
| Location | 7,684,810 – 7,684,900 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.49349 |
| G+C content | 0.45669 |
| Mean single sequence MFE | -17.55 |
| Consensus MFE | -6.19 |
| Energy contribution | -6.80 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7684810 90 + 21146708 AACCCACCACAUGCCCCA-UAGCUAUCCGAUUAUCUGUGAGAUAAUCGA---GUCCAGACCCAUCUCAGACUCAUAACUGUAGCACACUCCAUA--- ..................-..(((((..(((((((.....)))))))((---(((.(((....)))..)))))......)))))..........--- ( -19.70, z-score = -3.62, R) >droAna3.scaffold_13266 6663845 97 - 19884421 AACCCACCAUGUGACCCACUCCCUGUCUGAUUAUCUGUGAGAUAAUCGAUAGGUAGAGACCCAACUCAGACUCAUAACUAUAGCACACUCCACAAAA .........((((........((((((.(((((((.....)))))))))))))..(((......))).......................))))... ( -20.90, z-score = -2.38, R) >droEre2.scaffold_4845 4477440 77 - 22589142 ------------AACCCG-CCAC-AUCCGAUUAUCUGUGAGAUAAUCGA---GUCCAGACCCAUCCCAGACUCAUAACUGUAGCACACUCCAUG--- ------------.....(-(.((-(...(((((((.....)))))))((---(((..((....))...))))).....))).))..........--- ( -17.20, z-score = -3.55, R) >droYak2.chr2R 11870312 90 - 21139217 AACCCACCACAUGCCCCA-UAGCCAACCGAUUAUCUGUGAGAUAAUCGA---GUCCAGACCCAUCUCAGACUCAUAACUGUAGCACACUCCAUG--- ...........(((..((-.........(((((((.....)))))))((---(((.(((....)))..))))).....))..))).........--- ( -17.90, z-score = -3.19, R) >droSec1.super_1 5236840 90 + 14215200 AACUCACCUCAUGCCCCA-UAGCUAUCCGAUUAUCUGUGAGAUAAUCGA---GUCCAGACCCAUCUCAGACUCAUAACUGUAGCACACUCCAUG--- .........((((.....-..(((((..(((((((.....)))))))((---(((.(((....)))..)))))......)))))......))))--- ( -19.92, z-score = -2.97, R) >droSim1.chr2R 6212568 90 + 19596830 AACCCACCACAUGCCCCA-UAGCUAUCCGAUUAUCUGUGAGAUAAUCGA---GUCCAGACCCAUCUCAGACUCAUAACUGUAGCACACUCCAUG--- .........((((.....-..(((((..(((((((.....)))))))((---(((.(((....)))..)))))......)))))......))))--- ( -20.32, z-score = -3.71, R) >droWil1.scaffold_181009 3303029 75 - 3585778 -----------AUCCCUUUUUGUCCUUGCAUCAUCGAUAA--UCAUCAA---GUUGGGCUUGAA---AUAUUCAUAACUAUAGCACACUUCAAG--- -----------..(((..((((.......(((...)))..--....)))---)..)))((((((---.....................))))))--- ( -6.92, z-score = 0.61, R) >consensus AACCCACCACAUGCCCCA_UAGCUAUCCGAUUAUCUGUGAGAUAAUCGA___GUCCAGACCCAUCUCAGACUCAUAACUGUAGCACACUCCAUG___ ...........................((((((((.....))))))))................................................. ( -6.19 = -6.80 + 0.61)

| Location | 7,684,810 – 7,684,900 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.49349 |
| G+C content | 0.45669 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -10.10 |
| Energy contribution | -12.31 |
| Covariance contribution | 2.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 7684810 90 - 21146708 ---UAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGGUCUGGAC---UCGAUUAUCUCACAGAUAAUCGGAUAGCUA-UGGGGCAUGUGGUGGGUU ---(((.(.((((.(((.((((((((((((.(((....)))))))---))(((((((.....)))))))..)))))).-))).)))).).))).... ( -30.30, z-score = -2.54, R) >droAna3.scaffold_13266 6663845 97 + 19884421 UUUUGUGGAGUGUGCUAUAGUUAUGAGUCUGAGUUGGGUCUCUACCUAUCGAUUAUCUCACAGAUAAUCAGACAGGGAGUGGGUCACAUGGUGGGUU ..((((((......))))))....(((.((......)).))).((((((((((((((.....))))))).(((.........)))....))))))). ( -22.90, z-score = 0.81, R) >droEre2.scaffold_4845 4477440 77 + 22589142 ---CAUGGAGUGUGCUACAGUUAUGAGUCUGGGAUGGGUCUGGAC---UCGAUUAUCUCACAGAUAAUCGGAU-GUGG-CGGGUU------------ ---.......(.(((((((.....(((((..((.....))..)))---))(((((((.....)))))))...)-))))-)).)..------------ ( -27.40, z-score = -2.84, R) >droYak2.chr2R 11870312 90 + 21139217 ---CAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGGUCUGGAC---UCGAUUAUCUCACAGAUAAUCGGUUGGCUA-UGGGGCAUGUGGUGGGUU ---(((.(.((((.(((.(((((.((((((.(((....)))))))---))(((((((.....)))))))...))))).-))).)))).).))).... ( -30.60, z-score = -2.44, R) >droSec1.super_1 5236840 90 - 14215200 ---CAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGGUCUGGAC---UCGAUUAUCUCACAGAUAAUCGGAUAGCUA-UGGGGCAUGAGGUGAGUU ---(((...((((.(((.((((((((((((.(((....)))))))---))(((((((.....)))))))..)))))).-))).))))...))).... ( -31.20, z-score = -2.71, R) >droSim1.chr2R 6212568 90 - 19596830 ---CAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGGUCUGGAC---UCGAUUAUCUCACAGAUAAUCGGAUAGCUA-UGGGGCAUGUGGUGGGUU ---(((.(.((((.(((.((((((((((((.(((....)))))))---))(((((((.....)))))))..)))))).-))).)))).).))).... ( -32.40, z-score = -3.08, R) >droWil1.scaffold_181009 3303029 75 + 3585778 ---CUUGAAGUGUGCUAUAGUUAUGAAUAU---UUCAAGCCCAAC---UUGAUGA--UUAUCGAUGAUGCAAGGACAAAAAGGGAU----------- ---((((((((((..(((....))).))))---))))))(((..(---(((((.(--((...))).)).))))........)))..----------- ( -13.00, z-score = -0.38, R) >consensus ___CAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGGUCUGGAC___UCGAUUAUCUCACAGAUAAUCGGAUAGCUA_UGGGGCAUGUGGUGGGUU .........((((.(((..(((((((.((......)).))........(((((((((.....))))))))))))))...))).)))).......... (-10.10 = -12.31 + 2.21)

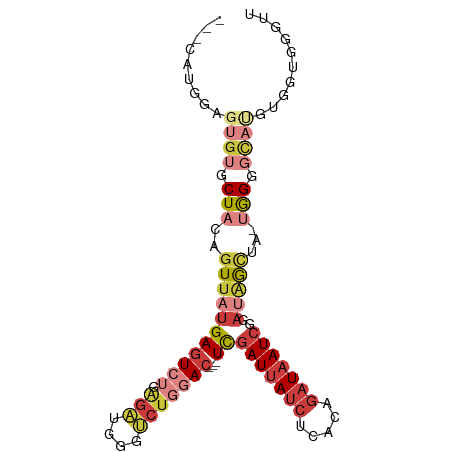
| Location | 7,684,865 – 7,684,968 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.93 |
| Shannon entropy | 0.55780 |
| G+C content | 0.44147 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -16.53 |
| Energy contribution | -18.71 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7684865 103 - 21146708 ------------UCUGGCUUUGG--AUUUGGUUUUUUCGAUUGGUUUCGGGCUCGUAAAAUGCACAGUUUAACGGUGUGCGGUAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGG ------------.......((((--(.........))))).(.(((((((((((((((...(((((.((((((.(....).)).)))).))))).....))))))))))))))).). ( -32.20, z-score = -2.88, R) >droEre2.scaffold_4845 4477482 101 + 22589142 ------------UCUGGCUUUGG--AUUUGG--UUUUGGAUUGCUUUUGGGCUCGUAAAAUGCUCAGUUUAACGGUGUGCGGCAUGGAGUGUGCUACAGUUAUGAGUCUGGGAUGGG ------------....((((..(--(.....--.))..))..))(((..(((((((((...((.((.(.....).)).))(((((.....)))))....)))))))))..))).... ( -24.40, z-score = 0.08, R) >droYak2.chr2R 11870367 114 + 21139217 UCUGUCUUUGGAUUUGGUUUUGA--UUUUGG-UUUUAGGAUUGCUUUUGGGCUCGUAAAAUGCACAGUUUAACGGUGUGCGGCAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGG .(((((((.(((..(((((((((--......-..)))))))))..)))((((((((((...(((((.(.....).)))))(((((.....)))))....))))))))))))))))). ( -31.10, z-score = -1.64, R) >droSec1.super_1 5236895 102 - 14215200 ------------UCUGGCUUUGG--AUUUGG-UUUUUCGAUUGGUUUCGGGCUCGUAAAAUGCACAGUUUAACGGUGUGCGGCAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGG ------------.......((((--(.....-...))))).(.(((((((((((((((...(((((.(.....).)))))(((((.....)))))....))))))))))))))).). ( -31.80, z-score = -2.29, R) >droSim1.chr2R 6212623 103 - 19596830 ------------UCUGGCUUUGG--AUUUGGUUUUUUCGAUUGCUUUCGGGCUCGUAAAAUGCACAGUUUAACGGUGUGCGGCAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGG ------------....((.((((--(.........)))))..))((((((((((((((...(((((.(.....).)))))(((((.....)))))....)))))))))))))).... ( -31.20, z-score = -1.76, R) >droWil1.scaffold_181009 3303072 101 + 3585778 ------UUUUCCACUGUUGUCGGCUCUUUUGUAGUCUCAUUUCUGUUGGUGAACAUAAAAUGCACAAUUUAAUGG-------CUUGAAGUGUGCUAUAGUUAUGAAUAUUUCAA--- ------..........(..(((((.....((......)).....)))))..).(((((...(((((.(((((...-------.))))).))))).....)))))..........--- ( -17.00, z-score = 0.30, R) >droVir3.scaffold_12875 15418151 79 + 20611582 -------------UUAUCUUUCU-GGUCUGG--UUCUGGUCUGGUCUGCGACUCGUAAAAUGCGCUAUUUAAUGG-------CUUGGAGUGUG--GCUAGUAUG------------- -------------........(.-.(.....--..)..).((((((.((.((((((.....))(((((...))))-------)...)))))))--)))))....------------- ( -14.90, z-score = 0.36, R) >consensus ____________UCUGGCUUUGG__AUUUGG_UUUUUCGAUUGCUUUCGGGCUCGUAAAAUGCACAGUUUAACGGUGUGCGGCAUGGAGUGUGCUACAGUUAUGAGUCUGAGAUGGG ............................................((((((((((((((...(((((.((((.((.....))...)))).))))).....)))))))))))))).... (-16.53 = -18.71 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:16 2011